Statistical structure of host-phage interactions
- PMID: 21709225
- PMCID: PMC3136311
- DOI: 10.1073/pnas.1101595108
Statistical structure of host-phage interactions
Abstract
Interactions between bacteria and the viruses that infect them (i.e., phages) have profound effects on biological processes, but despite their importance, little is known on the general structure of infection and resistance between most phages and bacteria. For example, are bacteria-phage communities characterized by complex patterns of overlapping exploitation networks, do they conform to a more ordered general pattern across all communities, or are they idiosyncratic and hard to predict from one ecosystem to the next? To answer these questions, we collect and present a detailed metaanalysis of 38 laboratory-verified studies of host-phage interactions representing almost 12,000 distinct experimental infection assays across a broad spectrum of taxa, habitat, and mode of selection. In so doing, we present evidence that currently available host-phage infection networks are statistically different from random networks and that they possess a characteristic nested structure. This nested structure is typified by the finding that hard to infect bacteria are infected by generalist phages (and not specialist phages) and that easy to infect bacteria are infected by generalist and specialist phages. Moreover, we find that currently available host-phage infection networks do not typically possess a modular structure. We explore possible underlying mechanisms and significance of the observed nested host-phage interaction structure. In addition, given that most of the available host-phage infection networks examined here are composed of taxa separated by short phylogenetic distances, we propose that the lack of modularity is a scale-dependent effect, and then, we describe experimental studies to test whether modular patterns exist at macroevolutionary scales.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
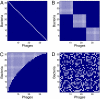





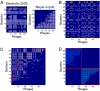

References
-
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. - PubMed
-
- Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. - PubMed
-
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: Making sense of genetic and ecological diversity. Science. 2009;323:741–746. - PubMed
-
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

