Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production
- PMID: 21711532
- PMCID: PMC3154180
- DOI: 10.1186/1752-0509-5-101
Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production
Abstract
Background: Ralstonia eutropha H16, found in both soil and water, is a Gram-negative lithoautotrophic bacterium that can utillize CO2 and H2 as its sources of carbon and energy in the absence of organic substrates. R. eutropha H16 can reach high cell densities either under lithoautotrophic or heterotrophic conditions, which makes it suitable for a number of biotechnological applications. It is the best known and most promising producer of polyhydroxyalkanoates (PHAs) from various carbon substrates and is an environmentally important bacterium that can degrade aromatic compounds. In order to make R. eutropha H16 a more efficient and robust biofactory, system-wide metabolic engineering to improve its metabolic performance is essential. Thus, it is necessary to analyze its metabolic characteristics systematically and optimize the entire metabolic network at systems level.
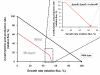
Results: We present the lithoautotrophic genome-scale metabolic model of R. eutropha H16 based on the annotated genome with biochemical and physiological information. The stoichiometic model, RehMBEL1391, is composed of 1391 reactions including 229 transport reactions and 1171 metabolites. Constraints-based flux analyses were performed to refine and validate the genome-scale metabolic model under environmental and genetic perturbations. First, the lithoautotrophic growth characteristics of R. eutropha H16 were investigated under varying feeding ratios of gas mixture. Second, the genome-scale metabolic model was used to design the strategies for the production of poly[R-(-)-3hydroxybutyrate] (PHB) under different pH values and carbon/nitrogen source uptake ratios. It was also used to analyze the metabolic characteristics of R. eutropha when the phosphofructokinase gene was expressed. Finally, in silico gene knockout simulations were performed to identify targets for metabolic engineering essential for the production of 2-methylcitric acid in R. eutropha H16.
Conclusion: The genome-scale metabolic model, RehMBEL1391, successfully represented metabolic characteristics of R. eutropha H16 at systems level. The reconstructed genome-scale metabolic model can be employed as an useful tool for understanding its metabolic capabilities, predicting its physiological consequences in response to various environmental and genetic changes, and developing strategies for systems metabolic engineering to improve its metabolic performance.
Figures


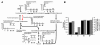
References
-
- Lee SY, Chang HN. Production of poly(3-hydroxybutyric acid) by recombinant Escherichia coli strains: genetic and fermentation studies. Can J Microbiol. 1995;41(Suppl 1):207–215. - PubMed
-
- Schlegel HG, Lafferty RM. Growth of Knallgas bacteria (Hydrogenomonas) using direct electrolysis of culture medium. Nature. 1965;205:308–309.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

