Mirtrons: microRNA biogenesis via splicing
- PMID: 21712066
- PMCID: PMC3185189
- DOI: 10.1016/j.biochi.2011.06.017
Mirtrons: microRNA biogenesis via splicing
Abstract
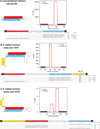
A well-defined mechanism governs the maturation of most microRNAs (miRNAs) in animals, via stepwise cleavage of precursor hairpin transcripts by the Drosha and Dicer RNase III enzymes. Recently, several alternative miRNA biogenesis pathways were elucidated, the most prominent of which substitutes Drosha cleavage with splicing. Such short hairpin introns are known as mirtrons, and their study has uncovered related pathways that combine splicing with other ribonucleolytic machinery to yield Dicer substrates for miRNA biogenesis. In this review, we consider the mechanisms of splicing-mediated miRNA biogenesis, computational strategies for mirtron discovery, and the evolutionary implications of the existence of multiple miRNA biogenesis pathways. Altogether, the features of mirtron pathways illustrate unexpected flexibility in combining RNA processing pathways, and highlight how multiple functions can be encoded by individual transcripts.
Copyright © 2011 Elsevier Masson SAS. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

