A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing
- PMID: 21712401
- PMCID: PMC3153973
- DOI: 10.1261/rna.2442211
A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing
Abstract
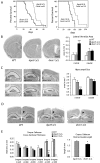
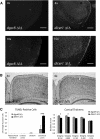
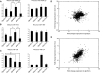
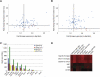
Noncanonical microRNAs (miRNAs) and endogenous small interfering RNAs (endo-siRNAs) are distinct subclasses of small RNAs that bypass the DGCR8/DROSHA Microprocessor but still require DICER1 for their biogenesis. What role, if any, they have in mammals remains unknown. To identify potential functional properties for these subclasses, we compared the phenotypes resulting from conditional deletion of Dgcr8 versus Dicer1 in post-mitotic neurons. The loss of Dicer1 resulted in an earlier lethality, more severe structural abnormalities, and increased apoptosis relative to that from Dgcr8 loss. Deep sequencing of small RNAs from the hippocampus and cortex of the conditional knockouts and control littermates identified multiple noncanonical microRNAs that were expressed at high levels in the brain relative to other tissues, including mirtrons and H/ACA snoRNA-derived small RNAs. In contrast, we found no evidence for endo-siRNAs in the brain. Taken together, our findings provide evidence for a diverse population of highly expressed noncanonical miRNAs that together are likely to play important functional roles in post-mitotic neurons.
Figures


References
-
- Babiarz JE, Blelloch R 2009. Small RNAs: their biogenesis, regulation and function in embryonic stem cells. In StemBook, ed. The Stem Cell Research Community. StemBook, Cambridge, MA - PubMed
-
- Brambilla P, Hardan A, di Nemi SU, Perez J, Soares JC, Barale F 2003. Brain anatomy and development in autism: review of structural MRI studies. Brain Res Bull 61: 557–569 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
