Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC)
- PMID: 21713444
- PMCID: PMC3219860
- DOI: 10.1007/s00203-011-0725-6
Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC)
Abstract
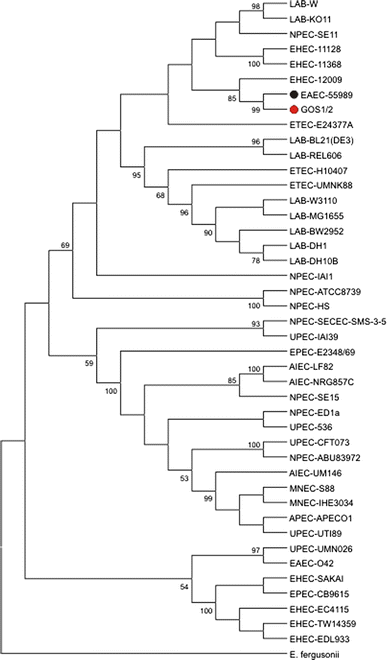
The genome sequences of two Escherichia coli O104:H4 strains derived from two different patients of the 2011 German E. coli outbreak were determined. The two analyzed strains were designated E. coli GOS1 and GOS2 (German outbreak strain). Both isolates comprise one chromosome of approximately 5.31 Mbp and two putative plasmids. Comparisons of the 5,217 (GOS1) and 5,224 (GOS2) predicted protein-encoding genes with various E. coli strains, and a multilocus sequence typing analysis revealed that the isolates were most similar to the entero-aggregative E. coli (EAEC) strain 55989. In addition, one of the putative plasmids of the outbreak strain is similar to pAA-type plasmids of EAEC strains, which contain aggregative adhesion fimbrial operons. The second putative plasmid harbors genes for extended-spectrum β-lactamases. This type of plasmid is widely distributed in pathogenic E. coli strains. A significant difference of the E. coli GOS1 and GOS2 genomes to those of EAEC strains is the presence of a prophage encoding the Shiga toxin, which is characteristic for enterohemorrhagic E. coli (EHEC) strains. The unique combination of genomic features of the German outbreak strain, containing characteristics from pathotypes EAEC and EHEC, suggested that it represents a new pathotype Entero-Aggregative-Haemorrhagic E scherichia c oli (EAHEC).
Figures




References
-
- Bernier C, Gounon P, Le Bouguenec C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF encoding operon family. Infect Immun. 2002;70:4302–4311. doi: 10.1128/IAI.70.8.4302-4311.2002. - DOI - PMC - PubMed
-
- Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J Clin Microbiol. 2005;43:452–454. doi: 10.1128/JCM.43.1.452-454.2005. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

