A hypomorphic mutation in Lpin1 induces progressively improving neuropathy and lipodystrophy in the rat
- PMID: 21715287
- PMCID: PMC3143639
- DOI: 10.1074/jbc.M110.197947
A hypomorphic mutation in Lpin1 induces progressively improving neuropathy and lipodystrophy in the rat
Abstract
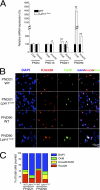
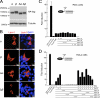
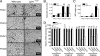
The Lpin1 gene encodes the phosphatidate phosphatase (PAP1) enzyme Lipin 1, which plays a critical role in lipid metabolism. In this study we describe the identification and characterization of a rat model with a mutated Lpin1 gene (Lpin1(1Hubr)), generated by N-ethyl-N-nitrosourea mutagenesis. Lpin1(1Hubr) rats are characterized by hindlimb paralysis and mild lipodystrophy that are detectable from the second postnatal week. Sequencing of Lpin1 identified a point mutation in the 5'-end splice site of intron 18 resulting in mis-splicing, a reading frameshift, and a premature stop codon. As this mutation does not induce nonsense-mediated decay, it allows the production of a truncated Lipin 1 protein lacking PAP1 activity. Lpin1(1Hubr) rats developed hypomyelination and mild lipodystrophy rather than the pronounced demyelination and adipocyte defects characteristic of Lpin1(fld/fld) mice, which carry a null allele for Lpin1. Furthermore, biochemical, histological, and molecular analyses revealed that these lesions improve in older Lpin1(1Hubr) rats as compared with young Lpin1(1Hubr) rats and Lpin1(fld/fld) mice. We observed activation of compensatory biochemical pathways substituting for missing PAP1 activity that, in combination with a possible non-enzymatic Lipin 1 function residing outside of its PAP1 domain, may contribute to the less severe phenotypes observed in Lpin1(1Hubr) rats as compared with Lpin1(fld/fld) mice. Although we are cautious in making a direct parallel between the presented rodent model and human disease, our data may provide new insight into the pathogenicity of recently identified human LPIN1 mutations.
Figures
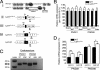




References
-
- Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Cell Metab. 4, 199–210 - PubMed
-
- Phan J., Reue K. (2005) Cell Metab. 1, 73–83 - PubMed
-
- Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007) J. Biol. Chem. 282, 3450–3457 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

