Transfection of plant mitochondria and in organello gene integration
- PMID: 21715377
- PMCID: PMC3177224
- DOI: 10.1093/nar/gkr517
Transfection of plant mitochondria and in organello gene integration
Abstract
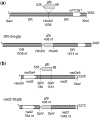
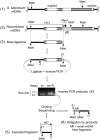
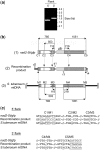
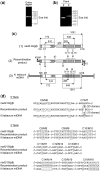
Investigation and manipulation of mitochondrial genetics in animal and plant cells remains restricted by the lack of an efficient in vivo transformation methodology. Mitochondrial transfection in whole cells and maintenance of the transfected DNA are main issues on this track. We showed earlier that isolated mitochondria from different organisms can import DNA. Exploiting this mechanism, we assessed the possibility to maintain exogenous DNA in plant organelles. Whereas homologous recombination is scarce in the higher plant nuclear compartment, recombination between large repeats generates the multipartite structure of the plant mitochondrial genome. These processes are under strict surveillance to avoid extensive genomic rearrangements. Nevertheless, following transfection of isolated organelles with constructs composed of a partial gfp gene flanked by fragments of mitochondrial DNA, we demonstrated in organello homologous recombination of the imported DNA with the resident DNA and integration of the reporter gene. Recombination yielded insertion of a continuous exogenous DNA fragment including the gfp sequence and at least 0.5 kb of flanking sequence on each side. According to our observations, transfection constructs carrying multiple sequences homologous to the mitochondrial DNA should be suitable and targeting of most regions in the organelle genome should be feasible, making the approach of general interest.
Figures



Similar articles
-
[Differential Expression of a Foreign Gene in Arabidopsis Mitochondria In Organelle].Mol Biol (Mosk). 2023 May-Jun;57(3):460-470. Mol Biol (Mosk). 2023. PMID: 37326049 Russian.
-
The plant mitochondrial genome: dynamics and maintenance.Biochimie. 2014 May;100:107-20. doi: 10.1016/j.biochi.2013.09.016. Epub 2013 Sep 26. Biochimie. 2014. PMID: 24075874 Review.
-
Elucidating genomic patterns and recombination events in plant cybrid mitochondria.Plant Mol Biol. 2019 Jul;100(4-5):433-450. doi: 10.1007/s11103-019-00869-z. Epub 2019 Apr 9. Plant Mol Biol. 2019. PMID: 30968307
-
DNA Import into Plant Mitochondria: Complex Approach for in organello and in vivo Studies.Biochemistry (Mosc). 2019 Jul;84(7):817-828. doi: 10.1134/S0006297919070113. Biochemistry (Mosc). 2019. PMID: 31509731
-
Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation.Annu Rev Plant Biol. 2017 Apr 28;68:225-252. doi: 10.1146/annurev-arplant-043015-112232. Epub 2017 Feb 9. Annu Rev Plant Biol. 2017. PMID: 28226235 Review.
Cited by
-
The Mosaic Mutants of Cucumber: A Method to Produce Knock-Downs of Mitochondrial Transcripts.G3 (Bethesda). 2015 Apr 14;5(6):1211-21. doi: 10.1534/g3.115.017053. G3 (Bethesda). 2015. PMID: 25873637 Free PMC article.
-
Gene delivery to mitochondria by targeting modified adenoassociated virus suppresses Leber's hereditary optic neuropathy in a mouse model.Proc Natl Acad Sci U S A. 2012 May 15;109(20):E1238-47. doi: 10.1073/pnas.1119577109. Epub 2012 Apr 20. Proc Natl Acad Sci U S A. 2012. PMID: 22523243 Free PMC article.
-
Polymer-coated carbon nanotube hybrids with functional peptides for gene delivery into plant mitochondria.Nat Commun. 2022 May 16;13(1):2417. doi: 10.1038/s41467-022-30185-y. Nat Commun. 2022. PMID: 35577779 Free PMC article.
-
Revisiting trends on mitochondrial mega-channels for the import of proteins and nucleic acids.J Bioenerg Biomembr. 2017 Feb;49(1):75-99. doi: 10.1007/s10863-016-9662-z. Epub 2016 May 5. J Bioenerg Biomembr. 2017. PMID: 27146409 Review.
-
Next-generation sequencing of mitochondrial targeted AAV transfer of human ND4 in mice.Mol Vis. 2013 Jul 14;19:1482-91. Print 2013. Mol Vis. 2013. PMID: 23869167 Free PMC article.
References
-
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797:113–128. - PubMed
-
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. - PubMed
-
- Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. - PubMed
-
- Holec S, Lange H, Canaday J, Gagliardi D. Coping with cryptic and defective transcripts in plant mitochondria. Biochim. Biophys. Acta. 2008;1779:566–573. - PubMed
-
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. - PubMed

