Adenovirus E1A directly targets the E2F/DP-1 complex
- PMID: 21715488
- PMCID: PMC3165805
- DOI: 10.1128/JVI.00539-11
Adenovirus E1A directly targets the E2F/DP-1 complex
Abstract
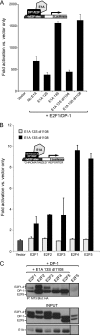
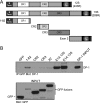
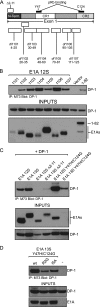
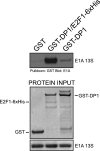
Deregulation of the cell cycle is of paramount importance during adenovirus infection. Adenovirus normally infects quiescent cells and must initiate the cell cycle in order to propagate itself. The pRb family of proteins controls entry into the cell cycle by interacting with and repressing transcriptional activation by the E2F transcription factors. The viral E1A proteins indirectly activate E2F-dependent transcription and cell cycle entry, in part, by interacting with pRb and family members to free the E2Fs. We report here that an E1A 13S isoform can unexpectedly activate E2F-responsive gene expression independently of binding to the pRb family of proteins. We demonstrate that E1A binds to E2F/DP-1 complexes through a direct interaction with DP-1. E1A appears to utilize this binding to recruit itself to E2F-regulated promoters, and this allows the E1A 13S protein, but not the E1A 12S protein, to activate transcription independently of interaction with pRb. Importantly, expression of E1A 13S, but not E1A 12S, led to significant enhancement of E2F4 occupancy of E2F sites of two E2F-regulated promoters. These observations identify a novel mechanism by which adenovirus deregulates the cell cycle and suggest that E1A 13S may selectively activate a subset of E2F-regulated cellular genes during infection.
Figures



References
-
- Avvakumov N., Kajon A. E., Hoeben R. C., Mymryk J. S. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329:477–492 - PubMed
-
- Bayley S. T., Mymryk J. S. 1994. Adenovirus E1A proteins and transformation. Int. J. Oncol. 5:425–444 - PubMed
-
- Berk A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 - PubMed
-
- Blais A., Dynlacht B. D. 2004. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 14:527–532 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

