Comparative evolution of GII.3 and GII.4 norovirus over a 31-year period
- PMID: 21715504
- PMCID: PMC3165818
- DOI: 10.1128/JVI.00472-11
Comparative evolution of GII.3 and GII.4 norovirus over a 31-year period
Abstract
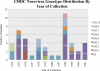
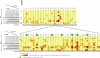
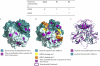
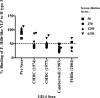
Noroviruses are the most common cause of epidemic gastroenteritis. Genotype II.3 is one of the most frequently detected noroviruses associated with sporadic infections. We studied the evolution of the major capsid gene from seven archival GII.3 noroviruses collected during a cross-sectional study at the Children's Hospital in Washington, DC, from 1975 through 1991, together with capsid sequence from 56 strains available in GenBank. Evolutionary analysis concluded that GII.3 viruses evolved at a rate of 4.16 × 10(-3) nucleotide substitutions/site/year (strict clock), which is similar to that described for the more prevalent GII.4 noroviruses. The analysis of the amino acid changes over the 31-year period found that GII.3 viruses evolve at a relatively steady state, maintaining 4% distance, and have a tendency to revert back to previously used residues while preserving the same carbohydrate binding profile. In contrast, GII.4 viruses demonstrate increasing rates of distance over time because of the continued integration of new amino acids and changing HBGA binding patterns. In GII.3 strains, seven sites acting under positive selection were predicted to be surface-exposed residues in the P2 domain, in contrast to GII.4 positively selected sites located primarily in the shell domain. Our study suggests that GII.3 noroviruses caused disease as early as 1975 and that they evolve via a specific pattern, responding to selective pressures induced by the host rather than presenting a nucleotide evolution rate lower than that of GII.4 noroviruses, as previously proposed. Understanding the evolutionary dynamics of prevalent noroviruses is relevant to the development of effective prevention and control strategies.
Figures



References
-
- Barreira D. M., et al. 2010. Viral load and genotypes of noroviruses in symptomatic and asymptomatic children in southeastern Brazil. J. Clin. Virol. 47:60–64 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

