Transcription factor plasmid binding modulates microtubule interactions and intracellular trafficking during gene transfer
- PMID: 21716302
- PMCID: PMC4150871
- DOI: 10.1038/gt.2011.96
Transcription factor plasmid binding modulates microtubule interactions and intracellular trafficking during gene transfer
Erratum in
- Gene Ther. 2012 Mar;19(3):354
Abstract
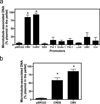
For non-viral gene delivery to be successful, plasmids must move through the cytoplasm to the nucleus in order to be transcribed. While the cytoskeletal meshwork acts as a barrier to plasmid DNA movement in the cytoplasm, the microtubule network is required for directed plasmid trafficking to the nucleus. We have shown previously that plasmid-microtubule interactions require cytoplasmic adapter proteins such as molecular motors, transcription factors (TFs) and importins. However, not all plasmid sequences support these interactions to allow movement to the nucleus. We now demonstrate that microtubule-DNA interactions can show sequence specificity with promoters containing binding sites for cyclic AMP response-element binding protein (CREB), including the cytomegalovirus immediate early promoter (CMV(iep)). Plasmids containing CREB-binding sites showed stringent interactions in an in vitro microtubule-binding assay. Using microinjection and real-time particle tracking, we show that the inclusion of TF binding sites within plasmids permits cytoplasmic trafficking of plasmids during gene transfer. We found that CREB-binding sites are bound by CREB in the cytoplasm during transfection, and allow for enhanced rates of movement and subsequent nuclear accumulation. Moreover, small interfering RNA knockdown of CREB prevented this enhanced trafficking. Therefore, TF binding sites within plasmids are necessary for interactions with microtubules and enhance movement to the nucleus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–208. - PubMed
-
- Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

