Baculovirus: molecular insights on their diversity and conservation
- PMID: 21716740
- PMCID: PMC3119482
- DOI: 10.4061/2011/379424
Baculovirus: molecular insights on their diversity and conservation
Abstract
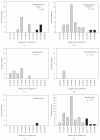
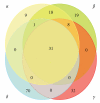
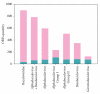
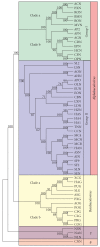
The Baculoviridae is a large group of insect viruses containing circular double-stranded DNA genomes of 80 to 180 kbp. In this study, genome sequences from 57 baculoviruses were analyzed to reevaluate the number and identity of core genes and to understand the distribution of the remaining coding sequences. Thirty one core genes with orthologs in all genomes were identified along with other 895 genes differing in their degrees of representation among reported genomes. Many of these latter genes are common to well-defined lineages, whereas others are unique to one or a few of the viruses. Phylogenetic analyses based on core gene sequences and the gene composition of the genomes supported the current division of the Baculoviridae into 4 genera: Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus.
Figures
Similar articles
-
On the classification and nomenclature of baculoviruses: a proposal for revision.Arch Virol. 2006 Jul;151(7):1257-66. doi: 10.1007/s00705-006-0763-6. Epub 2006 May 2. Arch Virol. 2006. PMID: 16648963 Review.
-
Codon usage in Alphabaculovirus and Betabaculovirus hosted by the same insect species is weak, selection dominated and exhibits no more similar patterns than expected.Infect Genet Evol. 2016 Oct;44:412-417. doi: 10.1016/j.meegid.2016.07.042. Epub 2016 Jul 30. Infect Genet Evol. 2016. PMID: 27484795 Free PMC article.
-
The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae.J Virol. 2012 Nov;86(22):12069-79. doi: 10.1128/JVI.01873-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933288 Free PMC article.
-
The Complete Genome of a New Betabaculovirus from Clostera anastomosis.PLoS One. 2015 Jul 13;10(7):e0132792. doi: 10.1371/journal.pone.0132792. eCollection 2015. PLoS One. 2015. PMID: 26168260 Free PMC article.
-
Baculovirus genomics.Curr Drug Targets. 2007 Oct;8(10):1051-68. doi: 10.2174/138945007782151333. Curr Drug Targets. 2007. PMID: 17979665 Review.
Cited by
-
Analysis of synonymous codon usage bias in helicase gene from Autographa californica multiple nucleopolyhedrovirus.Genes Genomics. 2018 Jul;40(7):767-780. doi: 10.1007/s13258-018-0689-x. Epub 2018 Apr 6. Genes Genomics. 2018. PMID: 29934813
-
The complete genome sequence of an alphabaculovirus from Spodoptera exempta, an agricultural pest of major economic significance in Africa.PLoS One. 2019 Feb 8;14(2):e0209937. doi: 10.1371/journal.pone.0209937. eCollection 2019. PLoS One. 2019. PMID: 30735528 Free PMC article.
-
Comparison of Chicken Immune Responses after Inoculation with H5 Avian Influenza Virus-like Particles Produced by Insect Cells or Pupae.J Vet Res. 2021 May 22;65(2):139-145. doi: 10.2478/jvetres-2021-0026. eCollection 2021 Jun. J Vet Res. 2021. PMID: 34250297 Free PMC article.
-
Using Next Generation Sequencing to Identify and Quantify the Genetic Composition of Resistance-Breaking Commercial Isolates of Cydia pomonella Granulovirus.Viruses. 2017 Sep 4;9(9):250. doi: 10.3390/v9090250. Viruses. 2017. PMID: 28869567 Free PMC article.
-
Advances in the Bioinformatics Knowledge of mRNA Polyadenylation in Baculovirus Genes.Viruses. 2020 Dec 6;12(12):1395. doi: 10.3390/v12121395. Viruses. 2020. PMID: 33291215 Free PMC article. Review.
References
-
- Jehle JA, Blissard GW, Bonning BC, et al. On the classification and nomenclature of baculoviruses: a proposal for revision. Archives of Virology. 2006;151(7):1257–1266. - PubMed
-
- Blissard GW, Rohrmann GF. Baculovirus diversity and molecular biology. Annual Review of Entomology. 1990;35(1):127–155. - PubMed
-
- Kozlov EA, Levitina TL, Gusak NM. The primary structure of baculovirus inclusion body proteins. Evolution and structure-function aspects. Current Topics in Microbiology and Immunology. 1986;131:135–164. - PubMed
-
- Rohrmann GF. Baculovirus structural proteins. Journal of General Virology. 1992;73(4):749–761. - PubMed
-
- Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. Journal of Virology. 2005;79(24):15258–15264. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
