Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex
- PMID: 21719644
- PMCID: PMC3601824
- DOI: 10.1126/science.1207125
Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex
Abstract
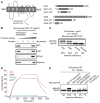
Tail-anchored (TA) proteins are involved in cellular processes including trafficking, degradation, and apoptosis. They contain a C-terminal membrane anchor and are posttranslationally delivered to the endoplasmic reticulum (ER) membrane by the Get3 adenosine triphosphatase interacting with the hetero-oligomeric Get1/2 receptor. We have determined crystal structures of Get3 in complex with the cytosolic domains of Get1 and Get2 in different functional states at 3.0, 3.2, and 4.6 angstrom resolution. The structural data, together with biochemical experiments, show that Get1 and Get2 use adjacent, partially overlapping binding sites and that both can bind simultaneously to Get3. Docking to the Get1/2 complex allows for conformational changes in Get3 that are required for TA protein insertion. These data suggest a molecular mechanism for nucleotide-regulated delivery of TA proteins.
Figures


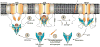
Comment in
-
It takes two to Get3.Structure. 2011 Oct 12;19(10):1353-5. doi: 10.1016/j.str.2011.10.001. Structure. 2011. PMID: 22000508
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

