Mechanism of RAD51-dependent DNA interstrand cross-link repair
- PMID: 21719678
- PMCID: PMC4068331
- DOI: 10.1126/science.1204258
Mechanism of RAD51-dependent DNA interstrand cross-link repair
Abstract
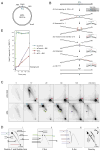
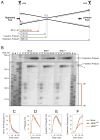
DNA interstrand cross-links (ICLs) are toxic DNA lesions whose repair in S phase of eukaryotic cells is incompletely understood. In Xenopus egg extracts, ICL repair is initiated when two replication forks converge on the lesion. Dual incisions then create a DNA double-strand break (DSB) in one sister chromatid, whereas lesion bypass restores the other sister. We report that the broken sister chromatid is repaired via RAD51-dependent strand invasion into the regenerated sister. Recombination acts downstream of FANCI-FANCD2, yet RAD51 binds ICL-stalled replication forks independently of FANCI-FANCD2 and before DSB formation. Our results elucidate the functional link between the Fanconi anemia pathway and the recombination machinery during ICL repair. In addition, they demonstrate the complete repair of a DSB via homologous recombination in vitro.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

