Physiological response to membrane protein overexpression in E. coli
- PMID: 21719796
- PMCID: PMC3205863
- DOI: 10.1074/mcp.M111.007930
Physiological response to membrane protein overexpression in E. coli
Abstract
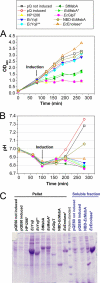
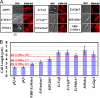
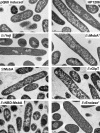
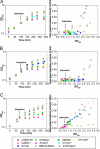
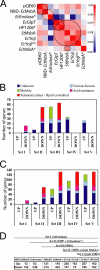
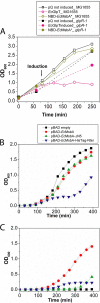
Overexpression represents a principal bottleneck in structural and functional studies of integral membrane proteins (IMPs). Although E. coli remains the leading organism for convenient and economical protein overexpression, many IMPs exhibit toxicity on induction in this host and give low yields of properly folded protein. Different mechanisms related to membrane biogenesis and IMP folding have been proposed to contribute to these problems, but there is limited understanding of the physical and physiological constraints on IMP overexpression and folding in vivo. Therefore, we used a variety of genetic, genomic, and microscopy techniques to characterize the physiological responses of Escherichia coli MG1655 cells to overexpression of a set of soluble proteins and IMPs, including constructs exhibiting different levels of toxicity and producing different levels of properly folded versus misfolded product on induction. Genetic marker studies coupled with transcriptomic results indicate only minor perturbations in many of the physiological systems implicated in previous studies of IMP biogenesis. Overexpression of either IMPs or soluble proteins tends to block execution of the standard stationary-phase transcriptional program, although these effects are consistently stronger for the IMPs included in our study. However, these perturbations are not an impediment to successful protein overexpression. We present evidence that, at least for the target proteins included in our study, there is no inherent obstacle to IMP overexpression in E. coli at moderate levels suitable for structural studies and that the biochemical and conformational properties of the proteins themselves are the major obstacles to success. Toxicity associated with target protein activity produces selective pressure leading to preferential growth of cells harboring expression-reducing and inactivating mutations, which can produce chemical heterogeneity in the target protein population, potentially contributing to the difficulties encountered in IMP crystallization.
Figures

References
-
- Gordon E., Horsefield R., Swarts H. G., de Pont J. J., Neutze R., Snijder A. (2008) Effective high-throughput overproduction of membrane proteins in Escherichia coli. Protein Expr. Purif. 62, 1–8 - PubMed
-
- Wagner S., Baars L., Ytterberg A. J., Klussmeier A., Wagner C. S., Nord O., Nygren P. A., van Wijk K. J., de Gier J. W. (2007) Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell Proteomics 6, 1527–1550 - PubMed
-
- Wang D. N., Safferling M., Lemieux M. J., Griffith H., Chen Y., Li X. D. (2003) Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim. Biophys. Acta 1610, 23–36 - PubMed
-
- White S. H. (2009) Biophysical dissection of membrane proteins. Nature 459, 344–346 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

