Structural and histone binding ability characterizations of human PWWP domains
- PMID: 21720545
- PMCID: PMC3119473
- DOI: 10.1371/journal.pone.0018919
Structural and histone binding ability characterizations of human PWWP domains
Abstract
Background: The PWWP domain was first identified as a structural motif of 100-130 amino acids in the WHSC1 protein and predicted to be a protein-protein interaction domain. It belongs to the Tudor domain 'Royal Family', which consists of Tudor, chromodomain, MBT and PWWP domains. While Tudor, chromodomain and MBT domains have long been known to bind methylated histones, PWWP was shown to exhibit histone binding ability only until recently.
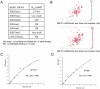
Methodology/principal findings: The PWWP domain has been shown to be a DNA binding domain, but sequence analysis and previous structural studies show that the PWWP domain exhibits significant similarity to other 'Royal Family' members, implying that the PWWP domain has the potential to bind histones. In order to further explore the function of the PWWP domain, we used the protein family approach to determine the crystal structures of the PWWP domains from seven different human proteins. Our fluorescence polarization binding studies show that PWWP domains have weak histone binding ability, which is also confirmed by our NMR titration experiments. Furthermore, we determined the crystal structures of the BRPF1 PWWP domain in complex with H3K36me3, and HDGF2 PWWP domain in complex with H3K79me3 and H4K20me3.
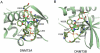
Conclusions: PWWP proteins constitute a new family of methyl lysine histone binders. The PWWP domain consists of three motifs: a canonical β-barrel core, an insertion motif between the second and third β-strands and a C-terminal α-helix bundle. Both the canonical β-barrel core and the insertion motif are directly involved in histone binding. The PWWP domain has been previously shown to be a DNA binding domain. Therefore, the PWWP domain exhibits dual functions: binding both DNA and methyllysine histones.
Enhanced version: This article can also be viewed as an enhanced version in which the text of the article is integrated with interactive 3D representations and animated transitions. Please note that a web plugin is required to access this enhanced functionality. Instructions for the installation and use of the web plugin are available in Text S1.
Conflict of interest statement
Figures





References
-
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. - PubMed
-
- Park J, Kim TY, Jung Y, Song SH, Kim SH, et al. DNA methyltransferase 3B mutant in ICF syndrome interacts non-covalently with SUMO-1. J Mol Med. 2008;86:1269–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

