New insights in plant immunity signaling activation
- PMID: 21723182
- PMCID: PMC3191233
- DOI: 10.1016/j.pbi.2011.05.005
New insights in plant immunity signaling activation
Abstract
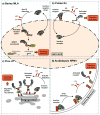
Plant disease resistance can be triggered by specific recognition of microbial effectors by plant nucleotide binding-leucine rich repeat (NB-LRR) receptors. Over the last few years, many efforts have greatly improved the understanding of effector and NB-LRR function, but have left a lot of questions as to how effector perception activates NB-LRR induction of defense signaling. This review describes exciting new findings showing similarities and differences in function of diverse plant NB-LRR proteins in terms of pathogen recognition and where and how resistance proteins are activated. Localization studies have shown that some NB-LRRs can activate signaling from the cytosol while others act in the nucleus. Also, the structural determination of two NB-LRR signaling domains demonstrated that receptor oligomerization is fundamental for the activation of resistance signaling.
Crown Copyright © 2011. Published by Elsevier Ltd. All rights reserved.
Figures
References
-
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. nature reviews genetics. 2010;11:539–548. - PubMed
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. - PubMed
-
- Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. - PubMed
-
- Inohara, Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

