A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri
- PMID: 21724612
- PMCID: PMC3185424
- DOI: 10.1093/nar/gkr521
A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri
Abstract
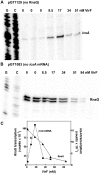
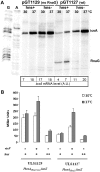
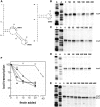
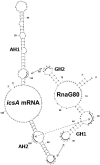
The icsA gene of Shigella encodes a structural protein involved in colonization of the intestinal mucosa by bacteria. This gene is expressed upon invasion of the host and is controlled by a complex regulatory circuit involving the nucleoid protein H-NS, the AraC-like transcriptional activator VirF, and a 450 nt antisense RNA (RnaG) acting as transcriptional attenuator. We investigated on the interplay of these factors at the molecular level. DNase I footprints reveal that both H-NS and VirF bind to a region including the icsA and RnaG promoters. H-NS is shown to repress icsA transcription at 30°C but not at 37°C, suggesting a significant involvement of this protein in the temperature-regulated expression of icsA. We also demonstrate that VirF directly stimulates icsA transcription and is able to alleviate H-NS repression in vitro. According to these results, icsA expression is derepressed in hns- background and overexpressed when VirF is provided in trans. Moreover, we find that RnaG-mediated transcription attenuation depends on 80 nt at its 5'-end, a stretch carrying the antisense region. Bases engaged in the initial contact leading to sense-antisense pairing have been identified using synthetic RNA and DNA oligonucleotides designed to rebuild and mutagenize the two stem-loop motifs of the antisense region.
Figures



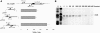
References
-
- Prosseda G, Falconi M, Nicoletti M, Casalino M, Micheli G, Colonna B. Histone-like proteins and the Shigella invasivity regulon. Res. Microbiol. 2002;153:461–468. - PubMed
-
- Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 2005;252:11–18. - PubMed
-
- Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 2003;50:45–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

