Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI
- PMID: 21724614
- PMCID: PMC3185434
- DOI: 10.1093/nar/gkr543
Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI
Abstract
A type IIG restriction endonuclease, RM.BpuSI from Bacillus pumilus, has been characterized and its X-ray crystal structure determined at 2.35Å resolution. The enzyme is comprised of an array of 5-folded domains that couple the enzyme's N-terminal endonuclease domain to its C-terminal target recognition and methylation activities. The REase domain contains a PD-x(15)-ExK motif, is closely superimposable against the FokI endonuclease domain, and coordinates a single metal ion. A helical bundle domain connects the endonuclease and methyltransferase (MTase) domains. The MTase domain is similar to the N6-adenine MTase M.TaqI, while the target recognition domain (TRD or specificity domain) resembles a truncated S subunit of Type I R-M system. A final structural domain, that may form additional DNA contacts, interrupts the TRD. DNA binding and cleavage must involve large movements of the endonuclease and TRD domains, that are probably tightly coordinated and coupled to target site methylation status.
Figures

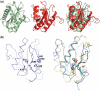
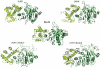


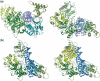
Similar articles
-
Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.J Mol Biol. 2008 Dec 12;384(2):489-502. doi: 10.1016/j.jmb.2008.09.033. Epub 2008 Sep 21. J Mol Biol. 2008. PMID: 18835275
-
The role of the methyltransferase domain of bifunctional restriction enzyme RM.BpuSI in cleavage activity.PLoS One. 2013 Nov 4;8(11):e80967. doi: 10.1371/journal.pone.0080967. eCollection 2013. PLoS One. 2013. PMID: 24224063 Free PMC article.
-
Functional analysis of MmeI from methanol utilizer Methylophilus methylotrophus, a subtype IIC restriction-modification enzyme related to type I enzymes.Appl Environ Microbiol. 2009 Jan;75(1):212-23. doi: 10.1128/AEM.01322-08. Epub 2008 Nov 7. Appl Environ Microbiol. 2009. PMID: 18997032 Free PMC article.
-
Type II restriction endonucleases: structure and mechanism.Cell Mol Life Sci. 2005 Mar;62(6):685-707. doi: 10.1007/s00018-004-4513-1. Cell Mol Life Sci. 2005. PMID: 15770420 Free PMC article. Review.
-
Structures, mechanisms, and kinetic advantages of the SgrAI filament forming mechanism.Crit Rev Biochem Mol Biol. 2024 Dec;59(6):363-401. doi: 10.1080/10409238.2024.2440315. Epub 2024 Dec 19. Crit Rev Biochem Mol Biol. 2024. PMID: 39699272 Review.
Cited by
-
Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria.Front Microbiol. 2023 Mar 22;14:1129437. doi: 10.3389/fmicb.2023.1129437. eCollection 2023. Front Microbiol. 2023. PMID: 37032876 Free PMC article. Review.
-
Resilience of biochemical activity in protein domains in the face of structural divergence.Curr Opin Struct Biol. 2014 Jun;26:92-103. doi: 10.1016/j.sbi.2014.05.008. Epub 2014 Jun 19. Curr Opin Struct Biol. 2014. PMID: 24952217 Free PMC article. Review.
-
Structural insights into DNA sequence recognition by Type ISP restriction-modification enzymes.Nucleic Acids Res. 2016 May 19;44(9):4396-408. doi: 10.1093/nar/gkw154. Epub 2016 Mar 14. Nucleic Acids Res. 2016. PMID: 26975655 Free PMC article.
-
Reappraisal of the DNA phosphorothioate modification machinery: uncovering neglected functional modalities and identification of new counter-invader defense systems.Nucleic Acids Res. 2024 Feb 9;52(3):1005-1026. doi: 10.1093/nar/gkad1213. Nucleic Acids Res. 2024. PMID: 38163645 Free PMC article.
-
Type I restriction enzymes and their relatives.Nucleic Acids Res. 2014 Jan;42(1):20-44. doi: 10.1093/nar/gkt847. Epub 2013 Sep 24. Nucleic Acids Res. 2014. PMID: 24068554 Free PMC article. Review.
References
-
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. - PubMed
-
- Dussoix D, Arber W. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 1962;5:37–49. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

