Involvement of the pleiotropic drug resistance response, protein kinase C signaling, and altered zinc homeostasis in resistance of Saccharomyces cerevisiae to diclofenac
- PMID: 21724882
- PMCID: PMC3165418
- DOI: 10.1128/AEM.00253-11
Involvement of the pleiotropic drug resistance response, protein kinase C signaling, and altered zinc homeostasis in resistance of Saccharomyces cerevisiae to diclofenac
Abstract
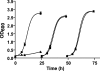
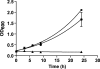
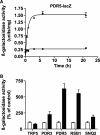
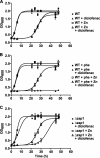
Diclofenac is a widely used analgesic drug that can cause serious adverse drug reactions. We used Saccharomyces cerevisiae as a model eukaryote with which to elucidate the molecular mechanisms of diclofenac toxicity and resistance. Although most yeast cells died during the initial diclofenac treatment, some survived and started growing again. Microarray analysis of the adapted cells identified three major processes involved in diclofenac detoxification and tolerance. In particular, pleiotropic drug resistance (PDR) genes and genes under the control of Rlm1p, a transcription factor in the protein kinase C (PKC) pathway, were upregulated in diclofenac-adapted cells. We tested if these processes or pathways were directly involved in diclofenac toxicity or resistance. Of the pleiotropic drug resistance gene products, the multidrug transporter Pdr5p was crucially important for diclofenac tolerance. Furthermore, deletion of components of the cell wall stress-responsive PKC pathway increased diclofenac toxicity, whereas incubation of cells with the cell wall stressor calcofluor white before the addition of diclofenac decreased its toxicity. Also, diclofenac induced flocculation, which might trigger the cell wall alterations. Genes involved in ribosome biogenesis and rRNA processing were downregulated, as were zinc-responsive genes. Paradoxically, deletion of the zinc-responsive transcription factor Zap1p or addition of the zinc chelator 1,10-phenanthroline significantly increased diclofenac toxicity, establishing a regulatory role for zinc in diclofenac resistance. In conclusion, we have identified three new pathways involved in diclofenac tolerance in yeast, namely, Pdr5p as the main contributor to the PDR response, cell wall signaling via the PKC pathway, and zinc homeostasis, regulated by Zap1p.
Figures

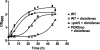
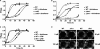
References
-
- Abou-Mohamed G., el-Kashef H. A., Salem H. A., Elmazar M. M. 1995. Effect of zinc on the anti-inflammatory and ulcerogenic activities of indometacin and diclofenac. Pharmacology 50:266–272 - PubMed
-
- Carvajal E., van den Hazel H. B., Cybularz-Kolaczkowska A., Balzi E., Goffeau A. 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256:406–415 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

