Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses
- PMID: 21724892
- PMCID: PMC3165258
- DOI: 10.1128/AEM.05017-11
Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses
Abstract
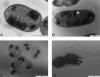
Sphagnum peatlands are important ecosystems in the methane cycle. Methane-oxidizing bacteria in these ecosystems serve as a methane filter and limit methane emissions. Yet little is known about the diversity and identity of the methanotrophs present in and on Sphagnum mosses of peatlands, and only a few isolates are known. The methanotrophic community in Sphagnum mosses, originating from a Dutch peat bog, was investigated using a pmoA microarray. A high biodiversity of both gamma- and alphaproteobacterial methanotrophs was found. With Sphagnum mosses as the inoculum, alpha- and gammaproteobacterial acidophilic methanotrophs were isolated using established and newly designed media. The 16S rRNA, pmoA, pxmA, and mmoX gene sequences showed that the alphaproteobacterial isolates belonged to the Methylocystis and Methylosinus genera. The Methylosinus species isolated are the first acid-tolerant members of this genus. Of the acidophilic gammaproteobacterial strains isolated, strain M5 was affiliated with the Methylomonas genus, and the other strain, M200, may represent a novel genus, most closely related to the genera Methylosoma and Methylovulum. So far, no acidophilic or acid-tolerant methanotrophs in the Gammaproteobacteria class are known. All strains showed the typical features of either type I or II methanotrophs and are, to the best of our knowledge, the first isolated (acidophilic or acid-tolerant) methanotrophs from Sphagnum mosses.
Figures






References
-
- Abell G. C. J., Stralis-Pavese N., Sessitsch A., Bodrossy L. 2009. Grazing affects methanotroph activity and diversity in an alpine meadow soil. Environ. Microbiol. Rep. 1:457–465 - PubMed
-
- Belova S. E., et al. 2011. Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3:36–46 - PubMed
-
- Belova S. E., Pankratov T. A., Dedysh S. N. 2006. Bacteria of the genus Burkholderia as a typical component of the microbial community of Sphagnum peat bogs. Microbiology 75:90–96 - PubMed
-
- Bodelier P. L. E., et al. 2009. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 3:606–617 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

