OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice
- PMID: 21725029
- PMCID: PMC3193001
- DOI: 10.1093/jxb/err144
OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice
Abstract
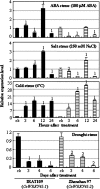
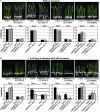
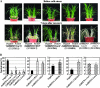
Although allelic diversity of genes has been shown to contribute to many phenotypic variations associated with different physiological processes in plants, information on allelic diversity of abiotic stress-responsive genes is limited. Here it is shown that the alleles OsWRKY45-1 and OsWRKY45-2 play different roles in abscisic acid (ABA) signalling and salt stress adaptation in rice. The two alleles had different transcriptional responses to ABA and salt stresses. OsWRKY45-1-overexpressing lines showed reduced ABA sensitivity, whereas OsWRKY45-1-knockout lines showed increased ABA sensitivity. OsWRKY45-1 transgenic plants showed no obvious difference from negative controls in response to salt stress. In contrast, OsWRKY45-2-overexpressing lines showed increased ABA sensitivity and reduced salt stress tolerance, and OsWRKY45-2-suppressing lines showed reduced ABA sensitivity and increased salt stress tolerance. OsWRKY45-1 and OsWRKY45-2 transgenic plants showed differential expression of a set of ABA- and abiotic stress-responsive genes, but they showed similar responses to cold and drought stresses. These results suggest that OsWRKY45-1 negatively and OsWRKY45-2 positively regulates ABA signalling and, in addition, OsWRKY45-2 but not OsWRKY45-1 negatively regulates rice response to salt stress. The different roles of the two alleles in ABA signalling and salt stress may be due to their transcriptional mediation of different signalling pathways.
Figures



References
-
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports. 2006;25:1263–1274. - PubMed
-
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. International Journal of Developmental Biology. 2005;49:717–732. - PubMed
-
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Molecular Biology. 2010;72:557–566. - PubMed
-
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. Antagonistic interaction between abscisic acid and jasmonates–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell. 2004;16:3460–3479. - PMC - PubMed
-
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009;69:473–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

