A large-scale method to measure absolute protein phosphorylation stoichiometries
- PMID: 21725298
- PMCID: PMC3146562
- DOI: 10.1038/nmeth.1636
A large-scale method to measure absolute protein phosphorylation stoichiometries
Abstract
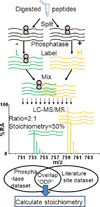
The functional role of protein phosphorylation is impacted by its fractional stoichiometry. Thus, a comprehensive strategy to study phosphorylation dynamics should include an assessment of site stoichiometry. Here we report an integrated method that relies on phosphatase treatment and stable-isotope labeling to determine absolute stoichiometries of protein phosphorylation on a large scale. This approach requires the measurement of only a single ratio relating phosphatase-treated and mock-treated samples. Using this strategy we determined stoichiometries for 5,033 phosphorylation sites in triplicate analyses from Saccharomyces cerevisiae growing through mid-log phase. We validated stoichiometries at ten sites that represented the full range of values obtained using synthetic phosphopeptides and found excellent agreement. Using bioinformatics, we characterized the biological properties associated with phosphorylation sites with vastly differing absolute stoichiometries.
Figures




Comment in
-
Phosphorylation sites of higher stoichiometry are more conserved.Nat Methods. 2012 Mar 27;9(4):317; author reply 318. doi: 10.1038/nmeth.1941. Nat Methods. 2012. PMID: 22453906 No abstract available.
References
-
- Macek B, Mann M, Olsen JV. Global and Site-Specific Quantitative Phosphoproteomics: Principles and Applications. Annu. Rev. Pharmacol. Toxicol. 2009;49:199–221. - PubMed
-
- Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9:1451–1468. - PubMed
-
- Boersema PJ, Mohammed S, Heck AJR. Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 2009;44:861–878. - PubMed
-
- Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nature Methods. 2007;4:798–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

