Small-molecule displacement of a cryptic degron causes conditional protein degradation
- PMID: 21725303
- PMCID: PMC3139708
- DOI: 10.1038/nchembio.598
Small-molecule displacement of a cryptic degron causes conditional protein degradation
Abstract
The ability to rapidly regulate the functions of specific proteins in living cells is a valuable tool for biological research. Here we describe a new technique by which the degradation of a specific protein is induced by a small molecule. A protein of interest is fused to a ligand-induced degradation (LID) domain, resulting in the expression of a stable and functional fusion protein. The LID domain is comprised of the FK506- and rapamycin-binding protein (FKBP) and a 19-amino-acid degron fused to the C terminus of FKBP. In the absence of the small molecule Shield-1, the degron is bound to the FKBP fusion protein and the protein is stable. When present, Shield-1 binds tightly to FKBP, displacing the degron and inducing rapid and processive degradation of the LID domain and any fused partner protein. Structure-function studies of the 19-residue peptide showed that a 4-amino-acid sequence within the peptide is responsible for degradation.
Figures
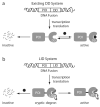




Comment in
-
Regulation through degradation.Nat Methods. 2011 Sep;8(9):711. doi: 10.1038/nmeth.1686. Nat Methods. 2011. PMID: 21985003
References
-
- Ryding ADS, Sharp MGF, Mullins JJ. Conditional transgenic technologies. J. Endocrinol. 2001;171:1–14. - PubMed
-
- Banaszynski LA, Wandless TJ. Conditional control of protein function. Chem. Biol. 2006;13:11–21. - PubMed
-
- Zhou P, Bogacki R, McReynolds L, Howley PM. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Mol. Cell. 2000;6:751–756. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

