A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition
- PMID: 21725307
- PMCID: PMC3145011
- DOI: 10.1038/ng.891
A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition
Abstract
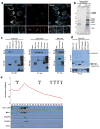
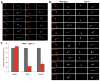
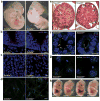
Mutations affecting ciliary components cause ciliopathies. As described here, we investigated Tectonic1 (Tctn1), a regulator of mouse Hedgehog signaling, and found that it is essential for ciliogenesis in some, but not all, tissues. Cell types that do not require Tctn1 for ciliogenesis require it to localize select membrane-associated proteins to the cilium, including Arl13b, AC3, Smoothened and Pkd2. Tctn1 forms a complex with multiple ciliopathy proteins associated with Meckel and Joubert syndromes, including Mks1, Tmem216, Tmem67, Cep290, B9d1, Tctn2 and Cc2d2a. Components of this complex co-localize at the transition zone, a region between the basal body and ciliary axoneme. Like Tctn1, loss of Tctn2, Tmem67 or Cc2d2a causes tissue-specific defects in ciliogenesis and ciliary membrane composition. Consistent with a shared function for complex components, we identified a mutation in TCTN1 that causes Joubert syndrome. Thus, a transition zone complex of Meckel and Joubert syndrome proteins regulates ciliary assembly and trafficking, suggesting that transition zone dysfunction is the cause of these ciliopathies.
Figures




Comment in
-
Transition zone proteins and cilia dynamics.Nat Genet. 2011 Jul 27;43(8):723-4. doi: 10.1038/ng.896. Nat Genet. 2011. PMID: 21792235 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- DK072301/DK/NIDDK NIH HHS/United States
- R25-GM56847/GM/NIGMS NIH HHS/United States
- R01 DK072301/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AR054396/AR/NIAMS NIH HHS/United States
- R01 DK068306/DK/NIDDK NIH HHS/United States
- RC4-K090917/RC/CCR NIH HHS/United States
- DK075972/DK/NIDDK NIH HHS/United States
- R25 GM056847/GM/NIGMS NIH HHS/United States
- R01 AR054396/AR/NIAMS NIH HHS/United States
- DK1068306/DK/NIDDK NIH HHS/United States
- HD04260/HD/NICHD NIH HHS/United States
- RC4 DK090917/DK/NIDDK NIH HHS/United States
- R01 HD042601/HD/NICHD NIH HHS/United States
- DK1069274/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

