Temperature-induced melting of double-stranded DNA in the absence and presence of covalently bonded antitumour drugs: insight from molecular dynamics simulations
- PMID: 21727089
- PMCID: PMC3185422
- DOI: 10.1093/nar/gkr512
Temperature-induced melting of double-stranded DNA in the absence and presence of covalently bonded antitumour drugs: insight from molecular dynamics simulations
Abstract
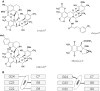
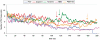
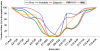
The difference in melting temperature of a double-stranded (ds) DNA molecule in the absence and presence of bound ligands can provide experimental information about the stabilization brought about by ligand binding. By simulating the dynamic behaviour of a duplex of sequence 5'-d(TAATAACGGATTATT)·5'-d(AATAATCCGTTATTA) in 0.1 M NaCl aqueous solution at 400 K, we have characterized in atomic detail its complete thermal denaturation profile in <200 ns. A striking asymmetry was observed on both sides of the central CGG triplet and the strand separation process was shown to be strongly affected by bonding in the minor groove of the prototypical interstrand crosslinker mitomycin C or the monofunctional tetrahydroisoquinolines trabectedin (Yondelis), Zalypsis and PM01183. Progressive helix unzipping was clearly interspersed with some reannealing events, which were most noticeable in the oligonucleotides containing the monoadducts, which maintained an average of 6 bp in the central region at the end of the simulations. These significant differences attest to the demonstrated ability of these drugs to stabilize dsDNA, stall replication and transcription forks, and recruit DNA repair proteins. This stabilization, quantified here in terms of undisrupted base pairs, supports the view that these monoadducts can functionally mimic a DNA interstrand crosslink.
Figures



References
-
- Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. - PubMed
-
- Stucki M, Stagljar I, Jónsson ZO, Hübscher U. A coordinated interplay: proteins with multiple functions in DNA replication, DNA repair, cell cycle/checkpoint control, and transcription. Prog. Nucleic Acid. Res. Mol. Biol. 2001;65:261–298. - PubMed
-
- Negri A, Marco E, García-Hernández V, Domingo A, Llamas-Saiz AL, Porto-Sandá S, Riguera R, Laine W, David-Cordonnier MH, Bailly C, et al. Antitumor activity, X-ray crystal structure, and DNA binding properties of thiocoraline A, a natural bis-intercalating thiodepsipeptide. J. Med. Chem. 2007;50:3322–3333. - PubMed
-
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996;355:13–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

