Predicting the functional impact of protein mutations: application to cancer genomics
- PMID: 21727090
- PMCID: PMC3177186
- DOI: 10.1093/nar/gkr407
Predicting the functional impact of protein mutations: application to cancer genomics
Abstract
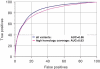
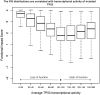
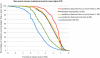
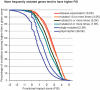
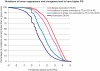
As large-scale re-sequencing of genomes reveals many protein mutations, especially in human cancer tissues, prediction of their likely functional impact becomes important practical goal. Here, we introduce a new functional impact score (FIS) for amino acid residue changes using evolutionary conservation patterns. The information in these patterns is derived from aligned families and sub-families of sequence homologs within and between species using combinatorial entropy formalism. The score performs well on a large set of human protein mutations in separating disease-associated variants (∼19 200), assumed to be strongly functional, from common polymorphisms (∼35 600), assumed to be weakly functional (area under the receiver operating characteristic curve of ∼0.86). In cancer, using recurrence, multiplicity and annotation for ∼10 000 mutations in the COSMIC database, the method does well in assigning higher scores to more likely functional mutations ('drivers'). To guide experimental prioritization, we report a list of about 1000 top human cancer genes frequently mutated in one or more cancer types ranked by likely functional impact; and, an additional 1000 candidate cancer genes with rare but likely functional mutations. In addition, we estimate that at least 5% of cancer-relevant mutations involve switch of function, rather than simply loss or gain of function.
Figures




References
-
- Ode H, Matsuyama S, Hata M, Neya S, Kakizawa J, Sugiura W, Hoshino T. Computational characterization of structural role of the non-active site mutation M36I of human immunodeficiency virus type 1 protease. J. Mol. Biol. 2007;370:598–607. - PubMed
-
- Lorch M, Mason JM, Sessions RB, Clarke AR. Effects of mutations on the thermodynamics of a protein folding reaction: implications for the mechanism of formation of the intermediate and transition states. Biochemistry. 2000;39:3480–3485. - PubMed
-
- Lorch M, Mason JM, Clarke AR, Parker MJ. Effects of core mutations on the folding of a beta-sheet protein: implications for backbone organization in the I-state. Biochemistry. 1999;38:1377–1385. - PubMed
-
- Alfalah M, Keiser M, Leeb T, Zimmer KP, Naim HY. Compound heterozygous mutations affect protein folding and function in patients with congenital sucrase-isomaltase deficiency. Gastroenterology. 2009;136:883–892. - PubMed
-
- Koukouritaki SB, Poch MT, Henderson MC, Siddens LK, Krueger SK, VanDyke JE, Williams DE, Pajewski NM, Wang T, Hines RN. Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants. J. Pharmacol. Exp. Ther. 2007;320:266–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

