Integration and analysis of genome-scale data from gliomas
- PMID: 21727940
- PMCID: PMC6980233
- DOI: 10.1038/nrneurol.2011.100
Integration and analysis of genome-scale data from gliomas
Abstract
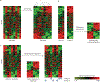
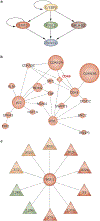
Primary brain tumors are a leading cause of cancer-related mortality among young adults and children. The most common primary malignant brain tumor, glioma, carries a median survival of only 14 months. Two major multi-institutional programs, the Glioma Molecular Diagnostic Initiative and The Cancer Genome Atlas, have pursued a comprehensive genomic characterization of a large number of clinical glioma samples using a variety of technologies to measure gene expression, chromosomal copy number alterations, somatic and germline mutations, DNA methylation, microRNA, and proteomic changes. Classification of gliomas on the basis of gene expression has revealed six major subtypes and provided insights into the underlying biology of each subtype. Integration of genome-wide data from different technologies has been used to identify many potential protein targets in this disease, while increasing the reliability and biological interpretability of results. Mapping genomic changes onto both known and inferred cellular networks represents the next level of analysis, and has yielded proteins with key roles in tumorigenesis. Ultimately, the information gained from these approaches will be used to create customized therapeutic regimens for each patient based on the unique genomic signature of the individual tumor. In this Review, we describe efforts to characterize gliomas using genomic data, and consider how insights gained from these analyses promise to increase understanding of the biological underpinnings of the disease and lead the way to new therapeutic strategies.
Figures


References
-
- Macdonald TJ et al. Expression profiling of medulloblastoma PDGFRA and the RAS/MAPK pathway. Nat. Genet 29, 143–152 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

