Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis
- PMID: 21730067
- PMCID: PMC3162405
- DOI: 10.1074/jbc.M111.247338
Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis
Abstract
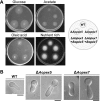
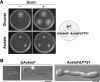
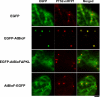
Among the eukaryotes only plants and a number of fungi are able to synthesize biotin. Although initial events leading to the biosynthesis of biotin remain largely unknown, the final steps are known to occur in the mitochondria. Here we deleted the Aopex5 and Aopex7 genes encoding the receptors for peroxisomal targeting signals PTS1 and PTS2, respectively, in the filamentous fungus Aspergillus oryzae. In addition to exhibiting defects in the peroxisomal targeting of either PTS1 or PTS2 proteins, the deletion strains also displayed growth defects on minimal medium containing oleic acid as the sole carbon source. Unexpectedly, these peroxisomal transport-deficient strains also exhibited growth defects on minimal medium containing glucose as the sole carbon source that were remediated by the addition of biotin and its precursors, including 7-keto-8-aminopelargonic acid (KAPA). Genome database searches in fungi and plants revealed that BioF protein/KAPA synthase, one of the biotin biosynthetic enzymes, has a PTS1 sequence at the C terminus. Fungal ΔbioF strains expressing the fungal and plant BioF proteins lacking PTS1 still exhibited growth defects in the absence of biotin, indicating that peroxisomal targeting of KAPA synthase is crucial for the biotin biosynthesis. Furthermore, in the plant Arabidopsis thaliana, AtBioF localized to the peroxisomes through recognition of its PTS1 sequence, suggesting involvement of peroxisomes in biotin biosynthesis in plants. Taken together we demonstrate a novel role for peroxisomes in biotin biosynthesis and suggest the presence of as yet unidentified peroxisomal proteins that function in the earlier steps of biotin biosynthesis.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

