Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR
- PMID: 21730181
- PMCID: PMC3145696
- DOI: 10.1073/pnas.1105848108
Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR
Abstract
Chromatin structure and function are regulated by numerous proteins through specific binding to nucleosomes. The structural basis of many of these interactions is unknown, as in the case of the high mobility group nucleosomal (HMGN) protein family that regulates various chromatin functions, including transcription. Here, we report the architecture of the HMGN2-nucleosome complex determined by a combination of methyl-transverse relaxation optimized nuclear magnetic resonance spectroscopy (methyl-TROSY) and mutational analysis. We found that HMGN2 binds to both the acidic patch in the H2A-H2B dimer and to nucleosomal DNA near the entry/exit point, "stapling" the histone core and the DNA. These results provide insight into how HMGNs regulate chromatin structure through interfering with the binding of linker histone H1 to the nucleosome as well as a structural basis of how phosphorylation induces dissociation of HMGNs from chromatin during mitosis. Importantly, our approach is generally applicable to the study of nucleosome-binding interactions in chromatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


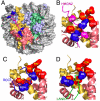
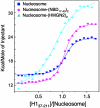
 , where Δδi is the difference in peak position between the free and bound states in ppm for atom i(i∈{Cγ1/2,Cδ1/2,Hγ1/2,Hδ1/2}), the 13C chemical shift weighting factor wi is set σH,i/σC,i (∼0.16–0.18), σi is the standard deviation of deposited chemical shifts in the Biological Magnetic Resonance Data Bank (41) for atom i, and the summation extends over the N methyl groups of each residue (i.e., N = 1 for Ile, and N = 2 for Leu/Val). (C) Methyl group peak intensity ratios, I(Mn2+)/I(Ca2+), of paramagnetic Mn2+-EDTA and diamagnetic Ca2+-EDTA spin-labeled HMGN2 at positions 19, 23, 38 and 44. Intensity ratios of the two individual methyl groups for Leu/Val were averaged. Errors (1 SD) are denoted by thin bars. Residues with average intensity ratio + 2 SD < 0.5 are labeled. (D), Structural summary of NMR CSP and paramagnetic spin labeling. Methyl groups with large chemical shift changes or decreases in peak intensity are labeled. Color coding as in Fig. 1. The sequence of the nucleosome-binding domain of HMGN2 and the position of the spin labels are shown (Left). Arg/Lys residues are highlighted in blue, and Ser24 and Ser28 are shown in magenta.
, where Δδi is the difference in peak position between the free and bound states in ppm for atom i(i∈{Cγ1/2,Cδ1/2,Hγ1/2,Hδ1/2}), the 13C chemical shift weighting factor wi is set σH,i/σC,i (∼0.16–0.18), σi is the standard deviation of deposited chemical shifts in the Biological Magnetic Resonance Data Bank (41) for atom i, and the summation extends over the N methyl groups of each residue (i.e., N = 1 for Ile, and N = 2 for Leu/Val). (C) Methyl group peak intensity ratios, I(Mn2+)/I(Ca2+), of paramagnetic Mn2+-EDTA and diamagnetic Ca2+-EDTA spin-labeled HMGN2 at positions 19, 23, 38 and 44. Intensity ratios of the two individual methyl groups for Leu/Val were averaged. Errors (1 SD) are denoted by thin bars. Residues with average intensity ratio + 2 SD < 0.5 are labeled. (D), Structural summary of NMR CSP and paramagnetic spin labeling. Methyl groups with large chemical shift changes or decreases in peak intensity are labeled. Color coding as in Fig. 1. The sequence of the nucleosome-binding domain of HMGN2 and the position of the spin labels are shown (Left). Arg/Lys residues are highlighted in blue, and Ser24 and Ser28 are shown in magenta.
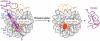
Comment in
-
Methyl fingerprinting of the nucleosome reveals the molecular mechanism of high-mobility group nucleosomal-2 (HMGN2) association.Proc Natl Acad Sci U S A. 2011 Jul 26;108(30):12189-90. doi: 10.1073/pnas.1109445108. Epub 2011 Jul 13. Proc Natl Acad Sci U S A. 2011. PMID: 21753078 Free PMC article. No abstract available.
References
-
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. - PubMed
-
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. - PubMed
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

