Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity
- PMID: 21730193
- PMCID: PMC3141992
- DOI: 10.1073/pnas.1105105108
Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity
Abstract
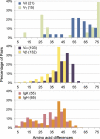
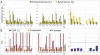
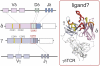
γδ T cells are an immunological enigma in that both their function in the immune response and the molecular mechanisms behind their activation remain unclear. These cells predominate in the epithelia and can be rapidly activated to provide an array of responses. However, no homologous γδ T-cell populations have been identified between humans and mice, and our understanding of what these cells recognize as ligands is limited. Here we take an alternative approach to understanding human γδ T-cell ligand recognition by studying the evolutionary forces that have shaped the V, D, and J gene segments that are used during somatic rearrangement to generate the γδ T-cell receptor. We find that distinctly different forces have shaped the γ and δ loci. The Vδ and Jδ genes are highly conserved, some even through to mouse. In contrast, the γ-locus is split: the Vγ9, Vγ10, and Vγ11 genes represent the conserved region of the Vγ gene locus whereas the remaining Vγ genes have been evolving rapidly, such that orthology throughout the primate lineage is unclear. We have also analyzed the coding versus silent substitutions between species within the V and J gene segments and find a preference for coding substitutions in the complementarity determining region loops of many of the V gene segments. Our results provide a different perspective on investigating human γδ T-cell recognition, demonstrating that diversification at particular γδ gene loci has been favored during primate evolution, suggesting adaptation of particular V domains to a changing ligand environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bonneville M, O'Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–478. - PubMed
-
- Nedellec S, Bonneville M, Scotet E. Human Vgamma9Vdelta2 T cells: from signals to functions. Semin Immunol. 2010;22:199–206. - PubMed
-
- Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. - PubMed
-
- Shin S, et al. Antigen recognition determinants of gammadelta T cell receptors. Science. 2005;308:252–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

