Generative embedding for model-based classification of fMRI data
- PMID: 21731479
- PMCID: PMC3121683
- DOI: 10.1371/journal.pcbi.1002079
Generative embedding for model-based classification of fMRI data
Abstract
Decoding models, such as those underlying multivariate classification algorithms, have been increasingly used to infer cognitive or clinical brain states from measures of brain activity obtained by functional magnetic resonance imaging (fMRI). The practicality of current classifiers, however, is restricted by two major challenges. First, due to the high data dimensionality and low sample size, algorithms struggle to separate informative from uninformative features, resulting in poor generalization performance. Second, popular discriminative methods such as support vector machines (SVMs) rarely afford mechanistic interpretability. In this paper, we address these issues by proposing a novel generative-embedding approach that incorporates neurobiologically interpretable generative models into discriminative classifiers. Our approach extends previous work on trial-by-trial classification for electrophysiological recordings to subject-by-subject classification for fMRI and offers two key advantages over conventional methods: it may provide more accurate predictions by exploiting discriminative information encoded in 'hidden' physiological quantities such as synaptic connection strengths; and it affords mechanistic interpretability of clinical classifications. Here, we introduce generative embedding for fMRI using a combination of dynamic causal models (DCMs) and SVMs. We propose a general procedure of DCM-based generative embedding for subject-wise classification, provide a concrete implementation, and suggest good-practice guidelines for unbiased application of generative embedding in the context of fMRI. We illustrate the utility of our approach by a clinical example in which we classify moderately aphasic patients and healthy controls using a DCM of thalamo-temporal regions during speech processing. Generative embedding achieves a near-perfect balanced classification accuracy of 98% and significantly outperforms conventional activation-based and correlation-based methods. This example demonstrates how disease states can be detected with very high accuracy and, at the same time, be interpreted mechanistically in terms of abnormalities in connectivity. We envisage that future applications of generative embedding may provide crucial advances in dissecting spectrum disorders into physiologically more well-defined subgroups.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 onto a probability distribution
onto a probability distribution  in a parametric family
in a parametric family  (see Sections ‘DCM for fMRI’ and ‘Model inversion’). In the second step (kernel construction), a kernel function
(see Sections ‘DCM for fMRI’ and ‘Model inversion’). In the second step (kernel construction), a kernel function  is defined that represents a similarity metric between any two fitted models
is defined that represents a similarity metric between any two fitted models  and
and  . This step can be split up into an initial mapping
. This step can be split up into an initial mapping  followed by a conventional kernel
followed by a conventional kernel  . The kernel implies a generative score space (or model-based feature space; see Section ‘Kernel construction’), which provides a comprehensive statistical representation of every subject. In this illustrative participant, the influence of region A on region B as well as the self-connection of region B were particularly strong. In the third step, a classifier is used to find a separating hyperplane between groups of subjects, based exclusively on their model-based representations (see Section ‘Classification’). When using a linear kernel, each feature corresponds to the coupling strength between two regions, which, in the fourth step, enables a mechanistic interpretation of feature weights in the context of the underlying model (see Section ‘Interpretation of the feature space’). Here, the influence of A on B and C were jointly most informative in distinguishing between groups. For a concrete implementation of this procedure, see Figure 2.
. The kernel implies a generative score space (or model-based feature space; see Section ‘Kernel construction’), which provides a comprehensive statistical representation of every subject. In this illustrative participant, the influence of region A on region B as well as the self-connection of region B were particularly strong. In the third step, a classifier is used to find a separating hyperplane between groups of subjects, based exclusively on their model-based representations (see Section ‘Classification’). When using a linear kernel, each feature corresponds to the coupling strength between two regions, which, in the fourth step, enables a mechanistic interpretation of feature weights in the context of the underlying model (see Section ‘Interpretation of the feature space’). Here, the influence of A on B and C were jointly most informative in distinguishing between groups. For a concrete implementation of this procedure, see Figure 2.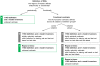

 , the model is specified using all subjects except
, the model is specified using all subjects except  . This yields a set of time series
. This yields a set of time series  for each subject
for each subject  . In step 2, the model is inverted independently for each subject, giving rise to a set of subject-specific posterior parameter means
. In step 2, the model is inverted independently for each subject, giving rise to a set of subject-specific posterior parameter means  . In step 3, these parameter estimates are used to train a classifier on all subjects except
. In step 3, these parameter estimates are used to train a classifier on all subjects except  and test it on subject
and test it on subject  , which yields a prediction about the class label of subject
, which yields a prediction about the class label of subject  . After having repeated these three steps for all
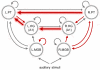
. After having repeated these three steps for all  , the set of predicted labels can be compared with the true labels, which allows us to estimate the algorithm's generalization performance. In addition, parameters that proved jointly discriminative can be interpreted in the context of the underlying generative model. The sequence of steps shown here corresponds to the procedure shown in Figure 2c and 2f, where it is contrasted with alternative procedures that are simpler but risk an optimistic bias in estimating generalization performance.
, the set of predicted labels can be compared with the true labels, which allows us to estimate the algorithm's generalization performance. In addition, parameters that proved jointly discriminative can be interpreted in the context of the underlying generative model. The sequence of steps shown here corresponds to the procedure shown in Figure 2c and 2f, where it is contrasted with alternative procedures that are simpler but risk an optimistic bias in estimating generalization performance.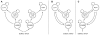




 -regularizer) was trained and tested in a leave-one-out cross-validation scheme, resulting in
-regularizer) was trained and tested in a leave-one-out cross-validation scheme, resulting in  subsets of selected features. The figure summarizes these subsets by visualizing how often each feature (printed along the y-axis) was selected across the
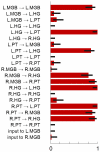
subsets of selected features. The figure summarizes these subsets by visualizing how often each feature (printed along the y-axis) was selected across the  repetitions (given as a fraction on the x-axis). Error bars represent central 95% posterior probability intervals of a Beta distribution with a flat prior over the interval [0, 1]. A group of 9 features was consistently found jointly informative for discriminating between aphasic patients and healthy controls (see main text). An additional figure showing which features were selected in each cross-validation fold can be found in the Supplementary Material (Figure S3). Crucially, since each feature corresponds to a model parameter that describes one particular interregional connection strength, the group of informative features can be directly related back to the underlying dynamic causal model (see highlighted connections in Figure 3).
repetitions (given as a fraction on the x-axis). Error bars represent central 95% posterior probability intervals of a Beta distribution with a flat prior over the interval [0, 1]. A group of 9 features was consistently found jointly informative for discriminating between aphasic patients and healthy controls (see main text). An additional figure showing which features were selected in each cross-validation fold can be found in the Supplementary Material (Figure S3). Crucially, since each feature corresponds to a model parameter that describes one particular interregional connection strength, the group of informative features can be directly related back to the underlying dynamic causal model (see highlighted connections in Figure 3).References
-
- Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, et al. Statistical parametric maps in functional imaging: A general linear approach. Hum Brain Mapp. 1995;2:189–210.
-
- Fu CH, Mourao-Miranda J, Costafreda SG, Khanna A, Marquand AF, et al. Pattern classification of sad facial processing: Toward the development of neurobiological markers in depression. Biol Psychiatry. 2008;63:656–662. - PubMed
-
- Shen H, Wang L, Liu Y, Hu D. Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. NeuroImage. 2010;49:3110–3121. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

