Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections
- PMID: 21731484
- PMCID: PMC3121881
- DOI: 10.1371/journal.ppat.1001346
Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections
Abstract
The obligate intracellular pathogen Chlamydia trachomatis is the most common cause of bacterial sexually transmitted diseases in the United States. In women C. trachomatis can establish persistent genital infections that lead to pelvic inflammatory disease and sterility. In contrast to natural infections in humans, experimentally induced infections with C. trachomatis in mice are rapidly cleared. The cytokine interferon-γ (IFNγ) plays a critical role in the clearance of C. trachomatis infections in mice. Because IFNγ induces an antimicrobial defense system in mice but not in humans that is composed of a large family of Immunity Related GTPases (IRGs), we questioned whether mice deficient in IRG immunity would develop persistent infections with C. trachomatis as observed in human patients. We found that IRG-deficient Irgm1/m3((-/-)) mice transiently develop high bacterial burden post intrauterine infection, but subsequently clear the infection more efficiently than wildtype mice. We show that the delayed but highly effective clearance of intrauterine C. trachomatis infections in Irgm1/m3((-/-)) mice is dependent on an exacerbated CD4(+) T cell response. These findings indicate that the absence of the predominant murine innate effector mechanism restricting C. trachomatis growth inside epithelial cells results in a compensatory adaptive immune response, which is at least in part driven by CD4(+) T cells and prevents the establishment of a persistent infection in mice.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

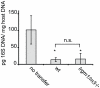

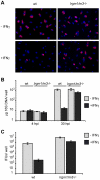

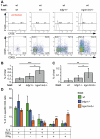
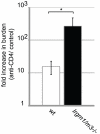

References
-
- Belland R, Ojcius DM, Byrne GI. Chlamydia. Nat Rev Microbiol. 2004;2:530–531. - PubMed
-
- World Health Organization. World Health Organization: Geneva; 2003. World Health Report, Shaping the future.
-
- Mpiga P, Ravaoarinoro M. Chlamydia trachomatis persistence: an update. Microbiol Res. 2006;161:9–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

