Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses
- PMID: 21731485
- PMCID: PMC3121877
- DOI: 10.1371/journal.ppat.1002074
Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses
Abstract
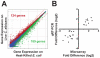
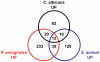
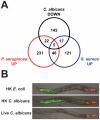
Candida albicans yeast cells are found in the intestine of most humans, yet this opportunist can invade host tissues and cause life-threatening infections in susceptible individuals. To better understand the host factors that underlie susceptibility to candidiasis, we developed a new model to study antifungal innate immunity. We demonstrate that the yeast form of C. albicans establishes an intestinal infection in Caenorhabditis elegans, whereas heat-killed yeast are avirulent. Genome-wide, transcription-profiling analysis of C. elegans infected with C. albicans yeast showed that exposure to C. albicans stimulated a rapid host response involving 313 genes (124 upregulated and 189 downregulated, ~1.6% of the genome) many of which encode antimicrobial, secreted or detoxification proteins. Interestingly, the host genes affected by C. albicans exposure overlapped only to a small extent with the distinct transcriptional responses to the pathogenic bacteria Pseudomonas aeruginosa or Staphylococcus aureus, indicating that there is a high degree of immune specificity toward different bacterial species and C. albicans. Furthermore, genes induced by P. aeruginosa and S. aureus were strongly over-represented among the genes downregulated during C. albicans infection, suggesting that in response to fungal pathogens, nematodes selectively repress the transcription of antibacterial immune effectors. A similar phenomenon is well known in the plant immune response, but has not been described previously in metazoans. Finally, 56% of the genes induced by live C. albicans were also upregulated by heat-killed yeast. These data suggest that a large part of the transcriptional response to C. albicans is mediated through "pattern recognition," an ancient immune surveillance mechanism able to detect conserved microbial molecules (so-called pathogen-associated molecular patterns or PAMPs). This study provides new information on the evolution and regulation of the innate immune response to divergent pathogens and demonstrates that nematodes selectively mount specific antifungal defenses at the expense of antibacterial responses.
Conflict of interest statement
RPW has served as a consultant for Optimer Pharmaceuticals, Inc. EM has received research support from and served on an advisory board for Astellas Pharamceuticals, Inc. The authors report no other potential conflicts of interest.
Figures





Similar articles
-
Thymol has antifungal activity against Candida albicans during infection and maintains the innate immune response required for function of the p38 MAPK signaling pathway in Caenorhabditis elegans.Immunol Res. 2016 Aug;64(4):1013-24. doi: 10.1007/s12026-016-8785-y. Immunol Res. 2016. PMID: 26783030
-
System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus.PLoS One. 2012;7(9):e44255. doi: 10.1371/journal.pone.0044255. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23028509 Free PMC article.
-
A Novel Virulence Phenotype Rapidly Assesses Candida Fungal Pathogenesis in Healthy and Immunocompromised Caenorhabditis elegans Hosts.mSphere. 2019 Apr 10;4(2):e00697-18. doi: 10.1128/mSphere.00697-18. mSphere. 2019. PMID: 30971447 Free PMC article.
-
Innate immune cell response upon Candida albicans infection.Virulence. 2016 Jul 3;7(5):512-26. doi: 10.1080/21505594.2016.1138201. Epub 2016 Apr 14. Virulence. 2016. PMID: 27078171 Free PMC article. Review.
-
Interplay between Candida albicans and the antimicrobial peptide armory.Eukaryot Cell. 2014 Aug;13(8):950-7. doi: 10.1128/EC.00093-14. Epub 2014 Jun 20. Eukaryot Cell. 2014. PMID: 24951441 Free PMC article. Review.
Cited by
-
Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection.PLoS Genet. 2012;8(6):e1002733. doi: 10.1371/journal.pgen.1002733. Epub 2012 Jun 14. PLoS Genet. 2012. PMID: 22719261 Free PMC article.
-
Antifungal efficacy during Candida krusei infection in non-conventional models correlates with the yeast in vitro susceptibility profile.PLoS One. 2013;8(3):e60047. doi: 10.1371/journal.pone.0060047. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555877 Free PMC article.
-
Caenorhabditis elegans, a model organism for investigating immunity.Appl Environ Microbiol. 2012 Apr;78(7):2075-81. doi: 10.1128/AEM.07486-11. Epub 2012 Jan 27. Appl Environ Microbiol. 2012. PMID: 22286994 Free PMC article. Review.
-
Multi-walled carbon nanotubes enhanced fungal colonization and suppressed innate immune response to fungal infection in nematodes.Toxicol Res (Camb). 2016 Jan 4;5(2):492-499. doi: 10.1039/c5tx00373c. eCollection 2016 Mar 1. Toxicol Res (Camb). 2016. PMID: 30090363 Free PMC article.
-
The contribution of Candida albicans vacuolar ATPase subunit V₁B, encoded by VMA2, to stress response, autophagy, and virulence is independent of environmental pH.Eukaryot Cell. 2014 Sep;13(9):1207-21. doi: 10.1128/EC.00135-14. Epub 2014 Jul 18. Eukaryot Cell. 2014. PMID: 25038082 Free PMC article.
References
-
- Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. - PubMed
-
- Leroy O, Gangneux JP, Montravers P, Mira JP, Gouin F, et al. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006). Crit Care Med. 2009;37:1612–1618. - PubMed
-
- Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37:1172–1177. - PubMed
-
- Leleu G, Aegerter P, Guidet B. Systemic candidiasis in intensive care units: a multicenter, matched-cohort study. J Crit Care. 2002;17:168–175. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

