Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses
- PMID: 21731485
- PMCID: PMC3121877
- DOI: 10.1371/journal.ppat.1002074
Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses
Abstract
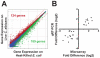
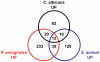
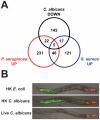
Candida albicans yeast cells are found in the intestine of most humans, yet this opportunist can invade host tissues and cause life-threatening infections in susceptible individuals. To better understand the host factors that underlie susceptibility to candidiasis, we developed a new model to study antifungal innate immunity. We demonstrate that the yeast form of C. albicans establishes an intestinal infection in Caenorhabditis elegans, whereas heat-killed yeast are avirulent. Genome-wide, transcription-profiling analysis of C. elegans infected with C. albicans yeast showed that exposure to C. albicans stimulated a rapid host response involving 313 genes (124 upregulated and 189 downregulated, ~1.6% of the genome) many of which encode antimicrobial, secreted or detoxification proteins. Interestingly, the host genes affected by C. albicans exposure overlapped only to a small extent with the distinct transcriptional responses to the pathogenic bacteria Pseudomonas aeruginosa or Staphylococcus aureus, indicating that there is a high degree of immune specificity toward different bacterial species and C. albicans. Furthermore, genes induced by P. aeruginosa and S. aureus were strongly over-represented among the genes downregulated during C. albicans infection, suggesting that in response to fungal pathogens, nematodes selectively repress the transcription of antibacterial immune effectors. A similar phenomenon is well known in the plant immune response, but has not been described previously in metazoans. Finally, 56% of the genes induced by live C. albicans were also upregulated by heat-killed yeast. These data suggest that a large part of the transcriptional response to C. albicans is mediated through "pattern recognition," an ancient immune surveillance mechanism able to detect conserved microbial molecules (so-called pathogen-associated molecular patterns or PAMPs). This study provides new information on the evolution and regulation of the innate immune response to divergent pathogens and demonstrates that nematodes selectively mount specific antifungal defenses at the expense of antibacterial responses.
Conflict of interest statement
RPW has served as a consultant for Optimer Pharmaceuticals, Inc. EM has received research support from and served on an advisory board for Astellas Pharamceuticals, Inc. The authors report no other potential conflicts of interest.
Figures





References
-
- Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. - PubMed
-
- Leroy O, Gangneux JP, Montravers P, Mira JP, Gouin F, et al. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006). Crit Care Med. 2009;37:1612–1618. - PubMed
-
- Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37:1172–1177. - PubMed
-
- Leleu G, Aegerter P, Guidet B. Systemic candidiasis in intensive care units: a multicenter, matched-cohort study. J Crit Care. 2002;17:168–175. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

