Evolutionary analysis of inter-farm transmission dynamics in a highly pathogenic avian influenza epidemic
- PMID: 21731491
- PMCID: PMC3121798
- DOI: 10.1371/journal.ppat.1002094
Evolutionary analysis of inter-farm transmission dynamics in a highly pathogenic avian influenza epidemic
Abstract
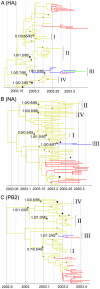
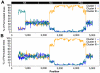
Phylogenetic studies have largely contributed to better understand the emergence, spread and evolution of highly pathogenic avian influenza during epidemics, but sampling of genetic data has never been detailed enough to allow mapping of the spatiotemporal spread of avian influenza viruses during a single epidemic. Here, we present genetic data of H7N7 viruses produced from 72% of the poultry farms infected during the 2003 epidemic in the Netherlands. We use phylogenetic analyses to unravel the pathways of virus transmission between farms and between infected areas. In addition, we investigated the evolutionary processes shaping viral genetic diversity, and assess how they could have affected our phylogenetic analyses. Our results show that the H7N7 virus was characterized by a high level of genetic diversity driven mainly by a high neutral substitution rate, purifying selection and limited positive selection. We also identified potential reassortment in the three genes that we have tested, but they had only a limited effect on the resolution of the inter-farm transmission network. Clonal sequencing analyses performed on six farm samples showed that at least one farm sample presented very complex virus diversity and was probably at the origin of chronological anomalies in the transmission network. However, most virus sequences could be grouped within clearly defined and chronologically sound clusters of infection and some likely transmission events between farms located 0.8-13 Km apart were identified. In addition, three farms were found as most likely source of virus introduction in distantly located new areas. These long distance transmission events were likely facilitated by human-mediated transport, underlining the need for strict enforcement of biosafety measures during outbreaks. This study shows that in-depth genetic analysis of virus outbreaks at multiple scales can provide critical information on virus transmission dynamics and can be used to increase our capacity to efficiently control epidemics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

