Model for biological communication in a nanofabricated cell-mimic driven by stochastic resonance
- PMID: 21731597
- PMCID: PMC3124924
- DOI: 10.1016/j.nancom.2011.03.001
Model for biological communication in a nanofabricated cell-mimic driven by stochastic resonance
Abstract
Cells offer natural examples of highly efficient networks of nanomachines. Accordingly, both intracellular and intercellular communication mechanisms in nature are looked to as a source of inspiration and instruction for engineered nanocommunication. Harnessing biological functionality in this manner requires an interdisciplinary approach that integrates systems biology, synthetic biology, and nanofabrication. Here, we present a model system that exemplifies the synergism between these realms of research. We propose a synthetic gene network for operation in a nanofabricated cell mimic array that propagates a biomolecular signal over long distances using the phenomenon of stochastic resonance. Our system consists of a bacterial quorum sensing signal molecule, a bistable genetic switch triggered by this signal, and an array of nanofabricated cell mimic wells that contain the genetic system. An optimal level of noise in the system helps to propagate a time-varying AHL signal over long distances through the array of mimics. This noise level is determined both by the system volume and by the parameters of the genetic network. Our proposed genetically driven stochastic resonance system serves as a testbed for exploring the potential harnessing of gene expression noise to aid in the transmission of a time-varying molecular signal.
Figures
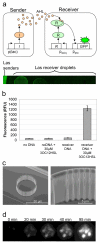
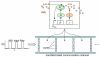
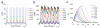
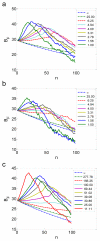
References
-
- Simpson ML, Cox CD, Peterson GD, Sayler GS. Engineering in the biological substrate: information processing in genetic circuits. Proceedings of the IEEE. 2004;92:848–863.
-
- Nakano T, Shuai J, Koujin T, Suda T, Hiraoka Y, Haraguchi T. Biological excitable media based on non-excitable cells and calcium signaling. Nano Communication Networks. 2010;1:43–49.
-
- Hiyama S, Moritani Y. Molecular communication: Harnessing biochemical materials to engineer biomimetic communication systems. Nano Communication Networks. 2010;1:20–30.
-
- Fernandes R, Roy V, Wu H-C, Bentley WE. Engineered biological nanofactories trigger quorum sensing response in targeted bacteria. Nat Nano. 2010;5:213–217. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
