Blood signature of pre-heart failure: a microarrays study
- PMID: 21731613
- PMCID: PMC3123284
- DOI: 10.1371/journal.pone.0020414
Blood signature of pre-heart failure: a microarrays study
Abstract
Background: The preclinical stage of systolic heart failure (HF), known as asymptomatic left ventricular dysfunction (ALVD), is diagnosed only by echocardiography, frequent in the general population and leads to a high risk of developing severe HF. Large scale screening for ALVD is a difficult task and represents a major unmet clinical challenge that requires the determination of ALVD biomarkers.
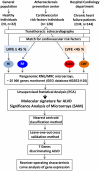
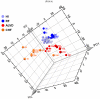
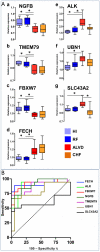
Methodology/principal findings: 294 individuals were screened by echocardiography. We identified 9 ALVD cases out of 128 subjects with cardiovascular risk factors. White blood cell gene expression profiling was performed using pangenomic microarrays. Data were analyzed using principal component analysis (PCA) and Significant Analysis of Microarrays (SAM). To build an ALVD classifier model, we used the nearest centroid classification method (NCCM) with the ClaNC software package. Classification performance was determined using the leave-one-out cross-validation method. Blood transcriptome analysis provided a specific molecular signature for ALVD which defined a model based on 7 genes capable of discriminating ALVD cases. Analysis of an ALVD patients validation group demonstrated that these genes are accurate diagnostic predictors for ALVD with 87% accuracy and 100% precision. Furthermore, Receiver Operating Characteristic curves of expression levels confirmed that 6 out of 7 genes discriminate for left ventricular dysfunction classification.
Conclusions/significance: These targets could serve to enhance the ability to efficiently detect ALVD by general care practitioners to facilitate preemptive initiation of medical treatment preventing the development of HF.
Conflict of interest statement
Figures
References
-
- Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, et al. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21–181. - PubMed
-
- Gottdiener JS, McClelland RL, Marshall R, Shemanski L, Furberg CD, et al. Outcome of congestive heart failure in elderly persons: influence of left ventricular systolic function. The Cardiovascular Health Study. Ann Intern Med. 2002;137:631–639. - PubMed
-
- Hunt SA, Abraham WT, Chin MH, Feldman AM, Francis GS, et al. 2009 focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:e391–479. - PubMed
-
- McMurray JV, McDonagh TA, Davie AP, Cleland JG, Francis CM, et al. Should we screen for asymptomatic left ventricular dysfunction to prevent heart failure? Eur Heart J. 1998;19:842–846. - PubMed
-
- Wang TJ, Levy D, Benjamin EJ, Vasan RS. The epidemiology of “asymptomatic” left ventricular systolic dysfunction: implications for screening. Ann Intern Med. 2003;138:907–916. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

