A comparative study of fragment screening methods on the p38α kinase: new methods, new insights
- PMID: 21732248
- PMCID: PMC3155752
- DOI: 10.1007/s10822-011-9454-9
A comparative study of fragment screening methods on the p38α kinase: new methods, new insights
Abstract
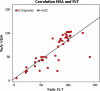
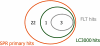
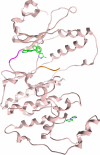
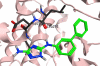
The stress-activated kinase p38α was used to evaluate a fragment-based drug discovery approach using the BioFocus fragment library. Compounds were screened by surface plasmon resonance (SPR) on a Biacore(™) T100 against p38α and two selectivity targets. A sub-set of our library was the focus of detailed follow-up analyses that included hit confirmation, affinity determination on 24 confirmed, selective hits and competition assays of these hits with respect to a known ATP binding site inhibitor. In addition, functional activity against p38α was assessed in a biochemical assay using a mobility shift platform (LC3000, Caliper LifeSciences). A selection of fragments was also evaluated using fluorescence lifetime (FLEXYTE(™)) and microscale thermophoresis (Nanotemper) technologies. A good correlation between the data for the different assays was found. Crystal structures were solved for four of the small molecules complexed to p38α. Interestingly, as determined both by X-ray analysis and SPR competition experiments, three of the complexes involved the fragment at the ATP binding site, while the fourth compound bound in a distal site that may offer potential as a novel drug target site. A first round of optimization around the remotely bound fragment has led to the identification of a series of triazole-containing compounds. This approach could form the basis for developing novel and active p38α inhibitors. More broadly, it illustrates the power of combining a range of biophysical and biochemical techniques to the discovery of fragments that facilitate the development of novel modulators of kinase and other drug targets.
Figures








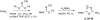
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

