An endothelial cell niche induces hepatic specification through dual repression of Wnt and Notch signaling
- PMID: 21732480
- PMCID: PMC3437666
- DOI: 10.1002/stem.576
An endothelial cell niche induces hepatic specification through dual repression of Wnt and Notch signaling
Abstract
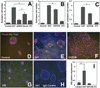
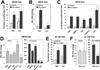
Complex cross-talk between endoderm and the microenvironment is an absolute requirement to orchestrate hepatic specification and expansion. In the mouse, the septum transversum and cardiac mesoderm, through secreted bone morphogenetic proteins (BMP) and fibroblast growth factors (FGF), respectively, instruct the adjacent ventral endoderm to become hepatic endoderm. Consecutively, endothelial cells promote expansion of the specified hepatic endoderm. By using a mouse reporter embryonic stem cell line, in which hCD4 and hCD25 were targeted to the Foxa2 and Foxa3 loci, we reconstituted an in vitro culture system in which committed endoderm cells coexpressing hCD4-Foxa2 and hCD25-Foxa3 were isolated and cocultured with endothelial cells in the presence of BMP4 and bFGF. In this culture setting, we provide mechanistic evidence that endothelial cells function not only to promote hepatic endoderm expansion but are also required at an earlier step for hepatic specification, at least in part through regulation of the Wnt and Notch pathways. Activation of Wnt and Notch by chemical or genetic approaches increases endoderm cell numbers but inhibits hepatic specification, and conversely, chemical inhibition of both pathways enhances hepatic specification and reduces proliferation. By using identical coculture conditions, we defined a similar dependence of endoderm harvested from embryos on endothelial cells to support their growth and hepatic specification. Our findings (1) confirm a conserved role of Wnt repression for mouse hepatic specification, (2) uncover a novel role for Notch repression in the hepatic fate decision, and (3) demonstrate that repression of Wnt and Notch signaling in hepatic endoderm is controlled by the endothelial cell niche.
Copyright © 2010 AlphaMed Press.
Conflict of interest statement
The authors indicate no potential conflict of interest.
Figures





Similar articles
-
Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications.Stem Cell Res. 2018 Jul;30:163-170. doi: 10.1016/j.scr.2018.06.004. Epub 2018 Jun 21. Stem Cell Res. 2018. PMID: 29936335
-
Anterior-Posterior Patterning of Definitive Endoderm Generated from Human Embryonic Stem Cells Depends on the Differential Signaling of Retinoic Acid, Wnt-, and BMP-Signaling.Stem Cells. 2016 Nov;34(11):2635-2647. doi: 10.1002/stem.2428. Epub 2016 Jul 4. Stem Cells. 2016. PMID: 27299363
-
FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast.Stem Cells Dev. 2012 Nov 1;21(16):2987-3000. doi: 10.1089/scd.2012.0099. Epub 2012 Aug 6. Stem Cells Dev. 2012. PMID: 22724507 Free PMC article.
-
Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells.Int J Dev Biol. 2013;57(1):1-12. doi: 10.1387/ijdb.120115ls. Int J Dev Biol. 2013. PMID: 23585347 Review.
-
Control of Blood Vessel Formation by Notch Signaling.Adv Exp Med Biol. 2018;1066:319-338. doi: 10.1007/978-3-319-89512-3_16. Adv Exp Med Biol. 2018. PMID: 30030834 Review.
Cited by
-
Notch signaling represses p63 expression in the developing surface ectoderm.Development. 2013 Sep;140(18):3777-86. doi: 10.1242/dev.093948. Epub 2013 Aug 7. Development. 2013. PMID: 23924630 Free PMC article.
-
Next-Generation Liver Medicine Using Organoid Models.Front Cell Dev Biol. 2019 Dec 20;7:345. doi: 10.3389/fcell.2019.00345. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31921856 Free PMC article. Review.
-
Generation of functional hepatic cells from pluripotent stem cells.J Stem Cell Res Ther. 2012 Aug 15;Suppl 10(8):1-7. doi: 10.4172/2157-7633.S10-008. J Stem Cell Res Ther. 2012. PMID: 25364624 Free PMC article.
-
Tissue engineering 2.0: guiding self-organization during pluripotent stem cell differentiation.Curr Opin Biotechnol. 2012 Oct;23(5):810-9. doi: 10.1016/j.copbio.2012.03.003. Epub 2012 Mar 21. Curr Opin Biotechnol. 2012. PMID: 22444525 Free PMC article. Review.
-
Orchestrating liver development.Development. 2015 Jun 15;142(12):2094-108. doi: 10.1242/dev.114215. Development. 2015. PMID: 26081571 Free PMC article. Review.
References
-
- Zaret KS. Liver specification and early morphogenesis. Mech Dev. 2000;92:83–88. - PubMed
-
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 18:175–189. - PubMed
-
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. - PubMed
-
- Jung J, Zheng M, Goldfarb M, et al. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

