MAL, but not MAL2, expression promotes the formation of cholesterol-dependent membrane domains that recruit apical proteins
- PMID: 21732912
- PMCID: PMC4390122
- DOI: 10.1042/BJ20110803
MAL, but not MAL2, expression promotes the formation of cholesterol-dependent membrane domains that recruit apical proteins
Abstract
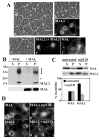
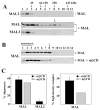
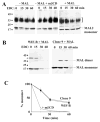
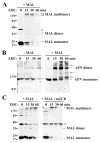
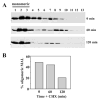
Our recent studies have been aimed at understanding the mechanisms regulating apical protein sorting in polarized epithelial cells. In particular, we have been investigating how lipid rafts serve to sort apical proteins in the biosynthetic pathway. The recent findings that lipid domains are too small or transient to host apically destined cargo have led to newer versions of the hypothesis that invoke proteins required for lipid domain coalescence and stabilization. MAL (myelin and lymphocyte protein) and its highly conserved family member, MAL2, have emerged as possible regulators of this process in the direct and indirect apical trafficking pathways respectively. To test this possibility, we took a biochemical approach. We determined that MAL, but not MAL2, self-associates, forms higher-order cholesterol-dependent complexes with apical proteins and promotes the formation of detergent-resistant membranes that recruit apical proteins. Such biochemical properties are consistent with a role for MAL in raft coalescence and stabilization. These findings also support a model whereby hydrophobic mismatch between the long membrane-spanning helices of MAL and the short-acyl-chain phospholipids in the Golgi drive formation of lipid domains rich in raft components that are characterized by a thicker hydrophobic core to alleviate mismatch.
Figures


Similar articles
-
Clustering and lateral concentration of raft lipids by the MAL protein.Mol Biol Cell. 2009 Aug;20(16):3751-62. doi: 10.1091/mbc.e09-02-0142. Epub 2009 Jun 24. Mol Biol Cell. 2009. PMID: 19553470 Free PMC article.
-
Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue.Histol Histopathol. 2004 Jul;19(3):925-33. doi: 10.14670/HH-19.925. Histol Histopathol. 2004. PMID: 15168355 Review.
-
Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells.Endocrinology. 2004 Feb;145(2):1011-6. doi: 10.1210/en.2003-0652. Epub 2003 Oct 23. Endocrinology. 2004. PMID: 14576188
-
MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells.J Cell Biol. 2002 Oct 14;159(1):37-44. doi: 10.1083/jcb.200206033. Epub 2002 Oct 7. J Cell Biol. 2002. PMID: 12370246 Free PMC article.
-
MAL, a proteolipid in glycosphingolipid enriched domains: functional implications in myelin and beyond.Prog Neurobiol. 2000 Apr;60(6):531-44. doi: 10.1016/s0301-0082(99)00039-8. Prog Neurobiol. 2000. PMID: 10739088 Review.
Cited by
-
CNS endothelial derived extracellular vesicles are biomarkers of active disease in multiple sclerosis.Fluids Barriers CNS. 2022 Feb 8;19(1):13. doi: 10.1186/s12987-021-00299-4. Fluids Barriers CNS. 2022. PMID: 35135557 Free PMC article.
-
MAL2 and rab17 selectively redistribute invadopodia proteins to laterally-induced protrusions in hepatocellular carcinoma cells.Mol Biol Cell. 2025 Mar 1;36(3):ar26. doi: 10.1091/mbc.E24-09-0400. Epub 2025 Jan 15. Mol Biol Cell. 2025. PMID: 39813085 Free PMC article.
-
The Molecular Architecture and Mode of Action of Clostridium perfringens ε-Toxin.Toxins (Basel). 2024 Apr 7;16(4):180. doi: 10.3390/toxins16040180. Toxins (Basel). 2024. PMID: 38668605 Free PMC article. Review.
-
Serine/threonine kinase 16 and MAL2 regulate constitutive secretion of soluble cargo in hepatic cells.Biochem J. 2014 Oct 15;463(2):201-13. doi: 10.1042/BJ20140468. Biochem J. 2014. PMID: 25084525 Free PMC article.
-
The cell biology of the hepatocyte: A membrane trafficking machine.J Cell Biol. 2019 Jul 1;218(7):2096-2112. doi: 10.1083/jcb.201903090. Epub 2019 Jun 14. J Cell Biol. 2019. PMID: 31201265 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

