A comparative analysis of DNA methylation across human embryonic stem cell lines
- PMID: 21733148
- PMCID: PMC3218824
- DOI: 10.1186/gb-2011-12-7-r62
A comparative analysis of DNA methylation across human embryonic stem cell lines
Abstract
Background: We performed a comparative analysis of the genome-wide DNA methylation profiles from three human embryonic stem cell (HESC) lines. It had previously been shown that HESC lines had significantly higher non-CG methylation than differentiated cells, and we therefore asked whether these sites were conserved across cell lines.
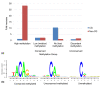
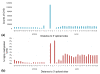
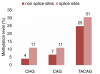
Results: We find that heavily methylated non-CG sites are strongly conserved, especially when found within the motif TACAG. They are enriched in splice sites and are more methylated than other non-CG sites in genes. We next studied the relationship between allele-specific expression and allele-specific methylation. By combining bisulfite sequencing and whole transcriptome shotgun sequencing (RNA-seq) data we identified 1,020 genes that show allele-specific expression, and 14% of CG sites genome-wide have allele-specific methylation. Finally, we asked whether the methylation state of transcription factor binding sites affects the binding of transcription factors. We identified variations in methylation levels at binding sites and found that for several transcription factors the correlation between the methylation at binding sites and gene expression is generally stronger than in the neighboring sequences.
Conclusions: These results suggest a possible but as yet unknown functional role for the highly methylated conserved non-CG sites in the regulation of HESCs. We also identified a novel set of genes that are likely transcriptionally regulated by methylation in an allele-specific manner. The analysis of transcription factor binding sites suggests that the methylation state of cis-regulatory elements impacts the ability of factors to bind and regulate transcription.
Figures