4D Flexible Atom-Pairs: An efficient probabilistic conformational space comparison for ligand-based virtual screening
- PMID: 21733172
- PMCID: PMC3156737
- DOI: 10.1186/1758-2946-3-23
4D Flexible Atom-Pairs: An efficient probabilistic conformational space comparison for ligand-based virtual screening
Abstract
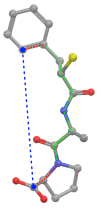
Background: The performance of 3D-based virtual screening similarity functions is affected by the applied conformations of compounds. Therefore, the results of 3D approaches are often less robust than 2D approaches. The application of 3D methods on multiple conformer data sets normally reduces this weakness, but entails a significant computational overhead. Therefore, we developed a special conformational space encoding by means of Gaussian mixture models and a similarity function that operates on these models. The application of a model-based encoding allows an efficient comparison of the conformational space of compounds.
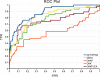
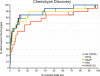
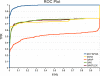
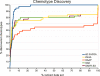
Results: Comparisons of our 4D flexible atom-pair approach with over 15 state-of-the-art 2D- and 3D-based virtual screening similarity functions on the 40 data sets of the Directory of Useful Decoys show a robust performance of our approach. Even 3D-based approaches that operate on multiple conformers yield inferior results. The 4D flexible atom-pair method achieves an averaged AUC value of 0.78 on the filtered Directory of Useful Decoys data sets. The best 2D- and 3D-based approaches of this study yield an AUC value of 0.74 and 0.72, respectively. As a result, the 4D flexible atom-pair approach achieves an average rank of 1.25 with respect to 15 other state-of-the-art similarity functions and four different evaluation metrics.
Conclusions: Our 4D method yields a robust performance on 40 pharmaceutically relevant targets. The conformational space encoding enables an efficient comparison of the conformational space. Therefore, the weakness of the 3D-based approaches on single conformations is circumvented. With over 100,000 similarity calculations on a single desktop CPU, the utilization of the 4D flexible atom-pair in real-world applications is feasible.
Figures

Similar articles
-
Probabilistic Modeling of Conformational Space for 3D Machine Learning Approaches.Mol Inform. 2010 May 17;29(5):441-55. doi: 10.1002/minf.201000036. Epub 2010 May 17. Mol Inform. 2010. PMID: 27463199
-
Stereoselective virtual screening of the ZINC database using atom pair 3D-fingerprints.J Cheminform. 2015 Feb 10;7:3. doi: 10.1186/s13321-014-0051-5. eCollection 2015. J Cheminform. 2015. PMID: 25750664 Free PMC article.
-
LS-align: an atom-level, flexible ligand structural alignment algorithm for high-throughput virtual screening.Bioinformatics. 2018 Jul 1;34(13):2209-2218. doi: 10.1093/bioinformatics/bty081. Bioinformatics. 2018. PMID: 29462237 Free PMC article.
-
Ligity: A Non-Superpositional, Knowledge-Based Approach to Virtual Screening.J Chem Inf Model. 2019 Jun 24;59(6):2600-2616. doi: 10.1021/acs.jcim.8b00779. Epub 2019 Jun 4. J Chem Inf Model. 2019. PMID: 31117509 Free PMC article.
-
Advances in the Development of Shape Similarity Methods and Their Application in Drug Discovery.Front Chem. 2018 Jul 25;6:315. doi: 10.3389/fchem.2018.00315. eCollection 2018. Front Chem. 2018. PMID: 30090808 Free PMC article. Review.
Cited by
-
Toward Quantum-Informed Atom Pairs.ACS Omega. 2024 Jan 26;9(5):5966-5971. doi: 10.1021/acsomega.3c09744. eCollection 2024 Feb 6. ACS Omega. 2024. PMID: 38343973 Free PMC article.
-
Conformational ensemble comparison for small molecules in drug discovery.J Comput Aided Mol Des. 2018 Aug;32(8):841-852. doi: 10.1007/s10822-018-0132-z. Epub 2018 Jul 9. J Comput Aided Mol Des. 2018. PMID: 29987709
-
Optimization and visualization of the edge weights in optimal assignment methods for virtual screening.BioData Min. 2013 Mar 26;6(1):7. doi: 10.1186/1756-0381-6-7. BioData Min. 2013. PMID: 23531368 Free PMC article.
-
Applying Machine Learning to Ultrafast Shape Recognition in Ligand-Based Virtual Screening.Front Pharmacol. 2020 Feb 19;10:1675. doi: 10.3389/fphar.2019.01675. eCollection 2019. Front Pharmacol. 2020. PMID: 32140104 Free PMC article.
References
-
- Varnek A, Tropsha A, (Eds) Chemoinformatics Approaches to Virtual Screening. Cambridge: The Royal Society of Chemistry; 2008.
LinkOut - more resources
Full Text Sources
Research Materials

