Stalled proteasomes are directly relieved by P97 recruitment
- PMID: 21733848
- PMCID: PMC3162386
- DOI: 10.1074/jbc.M111.240309
Stalled proteasomes are directly relieved by P97 recruitment
Abstract
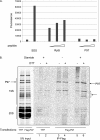
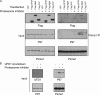
The 26 S proteasome is the eukaryotic protease responsible for the degradation of most cellular proteins. As such it accommodates the ability to function under diverse conditions that the cell may encounter. This function is supported by various adaptors that modulate various aspects in protein degradation, these include regulation of substrate delivery, deubiquitination, unfolding, and 20 S gate dilation. Here we show a new functional complex between the P97 and the proteasome that is assembled in response to proteasomal impairment. This entails P97 binding to the 26 S proteasome via the 19 S particle thereby forming an additional hexameric ATPase ring to relieve repression. P97-bound proteasomes showed selective binding toward the Npl4-ufd1 P97 co-factors, indicating a unique cellular role for P97 binding to proteasomes. P97-bound proteasomes display enhanced activity, showing a relief in proteolysis impairment. Our findings place P97 directly in non-ERAD proteasomal functions and establish a new checkpoint in UPS impairment. The ability to modulate proteasome activity and properly respond to protein misfolding, is of great importance in cellular regulation.
Figures





References
-
- Dobson C. M. (2003) Nature 426, 884–890 - PubMed
-
- Dobson C. M. (2003) Nat. Rev. Drug Discov. 2, 154–160 - PubMed
-
- Vendruscolo M., Zurdo J., MacPhee C. E., Dobson C. M. (2003) Philos. Transact. A Math Phys. Eng. Sci. 361, 1205–1222 - PubMed
-
- Macario A. J., Conway de Macario E. (2007) Front. Biosci. 12, 2588–2600 - PubMed
-
- Baumeister W., Lupas A. (1997) Curr. Opin. Struct. Biol. 7, 273–278 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

