Cancer biology and NuRD: a multifaceted chromatin remodelling complex
- PMID: 21734722
- PMCID: PMC4157524
- DOI: 10.1038/nrc3091
Cancer biology and NuRD: a multifaceted chromatin remodelling complex
Abstract
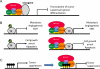
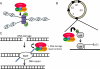
The nucleosome remodelling and histone deacetylase (NuRD; also known as Mi-2) complex regulates gene expression at the level of chromatin. The NuRD complex has been identified - using both genetic and molecular analyses - as a key determinant of differentiation in mouse embryonic stem cells and during development in various model systems. Similar to other chromatin remodellers, such as SWI/SNF and Polycomb complexes, NuRD has also been implicated in the regulation of transcriptional events that are integral to oncogenesis and cancer progression. Emerging molecular details regarding the recruitment of NuRD to specific loci during development, and the modulation of these events in cancer, are used to illustrate how the inappropriate localization of the complex could contribute to tumour biology.
Figures

References
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. - PubMed
-
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. - PubMed
-
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. - PubMed
-
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–6. - PubMed
-
- Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

