The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species
- PMID: 21734908
- PMCID: PMC3119406
- DOI: 10.3389/fmicb.2011.00105
The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species
Abstract
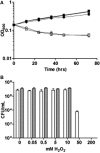
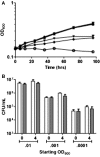
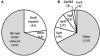
The bacteriostatic and bactericidal effects and the transcriptional response of Mycobacterium tuberculosis to representative oxidative and nitrosative stresses were investigated by growth and survival studies and whole genome expression analysis. The M. tuberculosis reaction to a range of hydrogen peroxide (H(2)O(2)) concentrations fell into three distinct categories: (1) low level exposure resulted in induction of a few highly sensitive H(2)O(2)-responsive genes, (2) intermediate exposure resulted in massive transcriptional changes without an effect on growth or survival, and (3) high exposure resulted in a muted transcriptional response and eventual death. M. tuberculosis appears highly resistant to DNA damage-dependent, mode-one killing caused by low millimolar levels of H(2)O(2) and only succumbs to overwhelming levels of oxidative stress observed in mode-two killing. Nitric oxide (NO) exposure initiated much the same transcriptional response as H(2)O(2). However, unlike H(2)O(2) exposure, NO exposure induced dormancy-related genes and caused dose-dependent bacteriostatic activity without killing. Included in the large shared response to H(2)O(2) and NO was the induction of genes encoding iron-sulfur cluster repair functions including iron acquisition. Stress regulons controlled by IdeR, Sigma H, Sigma E, and FurA comprised a large portion of the response to both stresses. Expression of several oxidative stress defense genes was constitutive, or increased moderately from an already elevated constitutive level, suggesting that bacilli are continually primed for oxidative stress defense.
Keywords: Mycobacterium tuberculosis; hydrogen peroxide; microarray; nitric oxide; reactive nitrogen species; reactive oxygen species.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

