B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1
- PMID: 21735360
- PMCID: PMC3925327
- DOI: 10.1007/82_2011_139
B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1
Abstract
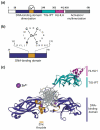
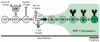
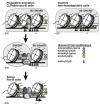
Early B cell factor 1 (EBF1) is a transcription factor that is critical for both B lymphopoiesis and B cell function. EBF1 is a requisite component of the B lymphocyte transcriptional network and is essential for B lineage specification. Recent studies revealed roles for EBF1 in B cell commitment. EBF1 binds its target genes via a DNA-binding domain including a unique 'zinc knuckle', which mediates a novel mode of DNA recognition. Chromatin immunoprecipitation of EBF1 in pro-B cells defined hundreds of new, as well as previously identified, target genes. Notably, expression of the pre-B cell receptor (pre-BCR), BCR and PI3K/Akt/mTOR signaling pathways is controlled by EBF1. In this review, we highlight these current developments and explore how EBF1 functions as a tissue-specific regulator of chromatin structure at B cell-specific genes.
Figures
References
-
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3 + lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. - PubMed
-
- Aravind L, Koonin E. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. - PubMed
-
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, van der Valk M, te Reile HPJ, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. - PubMed
-
- Bhalla S, Spaulding C, Brumbaugh RL, Zagort DE, Massari ME, Murre C, Kee BL. differential roles for the E2A activation domains in B lymphocytes and macrophages. J Immunol. 2008;180:1694–1703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

