Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization
- PMID: 21736719
- PMCID: PMC3142484
- DOI: 10.1186/1471-2180-11-161
Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization
Abstract
Background: Streptococcus suis is a zoonotic pathogen that causes infections in young piglets. S. suis is a heterogeneous species. Thirty-three different capsular serotypes have been described, that differ in virulence between as well as within serotypes.
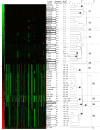
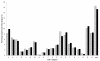
Results: In this study, the correlation between gene content, serotype, phenotype and virulence among 55 S. suis strains was studied using Comparative Genome Hybridization (CGH). Clustering of CGH data divided S. suis isolates into two clusters, A and B. Cluster A isolates could be discriminated from cluster B isolates based on the protein expression of extracellular factor (EF). Cluster A contained serotype 1 and 2 isolates that were correlated with virulence. Cluster B mainly contained serotype 7 and 9 isolates. Genetic similarity was observed between serotype 7 and serotype 2 isolates that do not express muramidase released protein (MRP) and EF (MRP⁻EF⁻), suggesting these isolates originated from a common founder. Profiles of 25 putative virulence-associated genes of S. suis were determined among the 55 isolates. Presence of all 25 genes was shown for cluster A isolates, whereas cluster B isolates lacked one or more putative virulence genes. Divergence of S. suis isolates was further studied based on the presence of 39 regions of difference. Conservation of genes was evaluated by the definition of a core genome that contained 78% of all ORFs in P1/7.
Conclusions: In conclusion, we show that CGH is a valuable method to study distribution of genes or gene clusters among isolates in detail, yielding information on genetic similarity, and virulence traits of S. suis isolates.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

