Generating libraries of iTol2-end insertions at BAC ends using loxP and lox511 Tn10 transposons
- PMID: 21736732
- PMCID: PMC3146455
- DOI: 10.1186/1471-2164-12-351
Generating libraries of iTol2-end insertions at BAC ends using loxP and lox511 Tn10 transposons
Abstract
Background: Bacterial Artificial Chromosomes (BACs) have been widely used as transgenes in vertebrate model systems such as mice and zebrafish, for a variety of studies. BAC transgenesis has been a powerful tool to study the function of the genome, and gene regulation by distal cis-regulatory elements. Recently, BAC transgenesis in both mice and zebrafish was further facilitated by development of the transposon-mediated method using the Tol2 element. Tol2 ends, in the inverted orientation and flanking a 1 kb spacer DNA (iTol2), were introduced into the BAC DNA within the bacterial host using recombination of homologous sequences. Here we describe experiments designed to determine if a simpler and more flexible system could modify BACs so that they would be suitable for transgenesis into zebrafish or mouse embryos using the Tol2 transposase.
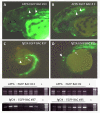
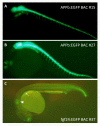
Results: A new technique was developed to introduce recognition sequences for the Tol2 transposase into BACs in E. coli using the Tn10 transposon vector system. We constructed pTnloxP-iTol2kan and pTnlox511-iTol2kan to introduce the loxP or lox511 site and iTol2 cassette, containing the Tol2 cis-sequences in the inverted orientation, into BACs that have loxP and lox511 sites flanking genomic DNA inserts by Tn10-mediated transposition. The procedure enables rapid generation of a large collection of BACs ready for transgenesis with the iTol2 cassette at the new end of a progressively truncated genomic insert via lox-Cre recombination. The iTol2 ends are efficiently recognized by the Tol2 transposase, and the BACs readily integrate into zebrafish chromosomes.
Conclusion: The new technology described here can rapidly introduce iTol2 ends at a BAC end of choice, and simultaneously generate a large collection of BACs with progressive deletions of the genomic DNA from that end in a single experiment. This procedure should be applicable to a wider variety of BACs containing lox sites flanking the genomic DNA insert, including those with sequence repeats. The libraries of iTol2 inserted BACs with truncations from an end should facilitate studies on the impact of distal cis-regulatory sequences on gene function, as well as standard BAC transgenesis with precisely trimmed genes in zebrafish or mouse embryos using Tol2 transposition.
Figures



References
-
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental Cell. 2004;7:133–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

