Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice
- PMID: 21737692
- PMCID: PMC3154886
- DOI: 10.1091/mbc.E11-01-0017
Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice
Abstract
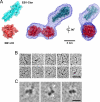
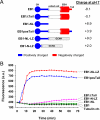
End-binding proteins (EBs) comprise a conserved family of microtubule plus end-tracking proteins. The concerted action of calponin homology (CH), linker, and C-terminal domains of EBs is important for their autonomous microtubule tip tracking, regulation of microtubule dynamics, and recruitment of numerous partners to microtubule ends. Here we report the detailed structural and biochemical analysis of mammalian EBs. Small-angle X-ray scattering, electron microscopy, and chemical cross-linking in combination with mass spectrometry indicate that EBs are elongated molecules with two interacting CH domains, an arrangement reminiscent of that seen in other microtubule- and actin-binding proteins. Removal of the negatively charged C-terminal tail did not affect the overall conformation of EBs; however, it increased the dwell times of EBs on the microtubule lattice in microtubule tip-tracking reconstitution experiments. An even more stable association with the microtubule lattice was observed when the entire negatively charged C-terminal domain of EBs was replaced by a neutral coiled-coil motif. In contrast, the interaction of EBs with growing microtubule tips was not significantly affected by these C-terminal domain mutations. Our data indicate that long-range electrostatic repulsive interactions between the C-terminus and the microtubule lattice drive the specificity of EBs for growing microtubule ends.
Figures




References
-
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. - PubMed
-
- Akhmanova A, Steinmetz HO. Microtubule end binding: EBs sense the guanine nucleotide state. Curr Biol. 2011;21:R283–R285. - PubMed
-
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

