Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1
- PMID: 21737788
- PMCID: PMC3163234
- DOI: 10.1161/CIRCRESAHA.111.241869
Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1
Abstract
Rationale: Oxidized palmitoyl arachidonyl phosphatidylcholine (Ox-PAPC) accumulates in atherosclerotic lesions, is proatherogenic, and influences the expression of more than 1000 genes in endothelial cells.
Objective: To elucidate the major pathways involved in Ox-PAPC action, we conducted a systems analysis of endothelial cell gene expression after exposure to Ox-PAPC.
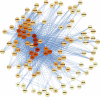
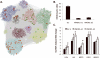
Methods and results: We used the variable responses of primary endothelial cells from 149 individuals exposed to Ox-PAPC to construct a network that consisted of 11 groups of genes, or modules. Modules were enriched for a broad range of Gene Ontology pathways, some of which have not been identified previously as major Ox-PAPC targets. Further validating our method of network construction, modules were consistent with relationships established by cell biology studies of Ox-PAPC effects on endothelial cells. This network provides novel hypotheses about molecular interactions, as well as candidate molecular regulators of inflammation and atherosclerosis. We validated several hypotheses based on network connections and genomic association. Our network analysis predicted that the hub gene CHAC1 (cation transport regulator homolog 1) was regulated by the ATF4 (activating transcription factor 4) arm of the unfolded protein response pathway, and here we showed that ATF4 directly activates an element in the CHAC1 promoter. We showed that variation in basal levels of heme oxygenase 1 (HMOX1) contribute to the response to Ox-PAPC, consistent with its position as a hub in our network. We also identified G-protein-coupled receptor 39 (GPR39) as a regulator of HMOX1 levels and showed that it modulates the promoter activity of HMOX1. We further showed that OKL38/OSGN1 (oxidative stress-induced growth inhibitor), the hub gene in the blue module, is a key regulator of both inflammatory and antiinflammatory molecules.
Conclusions: Our systems genetics approach has provided a broad view of the pathways involved in the response of endothelial cells to Ox-PAPC and also identified novel regulatory mechanisms.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

